INTRODUCTION
Peroxiredoxin (Prx) is a novel defined protein family which plays a critical role in reducing hydrogen peroxide with hydrogen derived from NAD(P)H[1-3]. The first member of this family was discovered as a 25 ku protein in yeast[4]. All Prx proteins contain a conserved cysteine residue in the N-terminal region that is the active site of catalysis. In mammal cells, there are six isoforms of peroxiredoxin, which can be devided into two subgroups, termed as 1-Cys Prx (VI) and 2-Cys Prx (I-V)[5,6]. The proteins of 2-Cys Prx subgroup contain another cysteine residue in the C-terminal portion of the molecule. Prx isoforms are distributed differently within cells and therefore with different functions. Prx I and II are localized to the cytosol[7,8], whereas Prx III has a mitochondrial-targeting signal and is localized in mitochondrion[9]. Prx IV is secreted out of cells because of its N-terminal sequence for secretion. Prx V is found in mitochondrion as long form of this protein, but it also exists in peroxisome as short form[10]. Same as Prx I and II, Prx VI is also found in cytosol[11].
Prx I, which belongs to 2-Cys subgroup, is also known as MSP23, PAG and NEEF-A[12-14]. Prx I is involved in redox regulation of the cell, such as reducing peroxides by reducing equivalents provided through the thioredoxin system but not from glutaredoxin. Prx I is found in abundance in the cytoplasm of cells as homodimers. Its cDNA was first cloned from mouse peritoneal macrophage encoding a protein of 23 ku. Now the antioxidative molecular mechanism of Prx I is elucidated by several researches on molecular structure has been performed. From the results of biochemical and physiological studies, Prx I has been implicated in a number of cellular functions, such as cell proliferation and differentiation, enhancement of the natural killer cell activity, and intracellular signaling, in addition to the antioxidant activity[15-17]. Further studies have shown that the expression of Prx I is increased in cancer cells, indicating it may be related to cancer development or progression[18]. We postulate that it may have an activity of anti-tumor development.
In our previous study, we found the expression of Prx I was up regulated by ionizing radiation in mouse intestinal epithelia[19]. This was coincident with the results of other researchers[20]. As a step to further study the biochemical functions of Prx I, we cloned the coding region of this gene and expressed it in IEC-6 cells. Our data showed that Prx I could be successfully expressed in this kind of cells.
MATERIALS AND METHODS
Materials
Male BALB/c mice, 58-62 d of age (weight 20-24 g), were purchased from Animal Center in Third Military Medical University, and housed in conventional cages with free access to drinking water and standard chow. Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum were purchased from Hyclone (Logan, USA). RNA PCR kit and T-vector were obtained from Takara (Dalian, China), and antibodies were from Santa Cruz (California, USA). Reagent for RNA isolation was from Roche Ltd. (Roche, USA). Plasmid pSG5 was a kindly gift from Dr. Chen Jian (Department of Biochemistry, Third Military Medical University). The specific primers were synthesized by Shanghai Shenyou Shengwu Jishu Corporation. Other chemicals not mentioned above were domestic made.
Isolation of total RNAs
Mouse intestinal epithelial cells were isolated as previously described[21]. After the cell pipette washed with cold PBS (0.01 mol/L, pH7.4), the cells were lysed with Tripure solution. All performs were according to the manufacturer’s instructions. The quality of isolated RNAs was checked by electrophoresis.
RT-PCR
The coding region of Prx I was amplified by RT-PCR with forward primer, 5’CGAATTCGTTCTCACGGCTCTTTCTGTTT3’ (P1) and backward primer, 5’AGGATCCTTCTGGCTGCTCAATGC TGC3’ (P2). One microgram of total RNAs was added into each reaction. After the reverse transcription was performed at 50 °C for 30 min, the amplification was carried out with one denaturing cycle of 3 min at 94 °C, then 28 cycles consisting of denaturation at 94 °C for 40 s, annealing at 58 °C for 40 s, and extension at 72 °C for 40 s, and one additional extension at 72 °C for 5 min. RT-PCR products were analyzed on 12 g/L agarose gel. To validate the transcription of recombinant plasmid, the mRNA of transfected IEC-6 cells was amplified by RT-PCR with primers P2 and P3 (5’AAAGCTGGATCGATCCTGAG3’). The procedure in details was performed according to the manufacturer’s instructions.
Construction of expression vector
The amplified DNA was separated by electrophorsis and purified from gels. Then 1µL of T-vector, 1µL of T4 DNA ligase and 2µL of 10 × buffer were added to the purified RT-PCR product, and incubated at 16 °C overnight. Competent bacteria of E. coli JM109 were transformed with the linkage product, and then spread on a LB agarose plate containing ampicillin and X-gal. The inserted DNA into T-vector of 3 random monoclonal colonies was sequenced, and the colony was termed as pT-Prx. To construct the recombinant expression vector, the EcoR I-BamH I fragment of pT-Prx was inserted into vector pSG5, which could transfect IEC-6 cells.
Cell culture and transfection
IEC-6 cells were cultivated at 37 °C in an atmosphere containing 100 mL/L CO2 in air. The culture medium comprised of DMEM containing 100 mL/L fetal calf serum, penicillin (100 U/mL), and streptomycin (100 µg/mL). Exponentially growing cells were transferred to 100 mL culture dishes and cultured for 24 h to yield about 40% confluency at the time of transfection. Before transfection, 30 µg of DOTAP was diluted with 70 µL of HBS solution (20 mmol/L HEPES, 150 mmol/L NaCl, pH7.4), and 5 µg of DNA was diluted to 0.1µg/µL with the same solution. The mixture of DNA and DOTAP was prepared by gently mixing them together and incubated for 15 min at room temperature. To prepare cells for transfection, IEC-6 cells were washed twice with serum-free medium. After the cells were incubated with the mixture of DNA and DOTAP in 2 mL of serum-free medium for 4 h, the cells were washed with serum-free medium, followed by further culture for 16 h. Transfection was repeated one more time. At the time of 24 h after the second transfection, the cells were collected for RNA and proteins isolation.
Immunoblotting
IEC-6 cells were collected and washed with ice-cold PBS (0.01 mol/L, pH7.4) after transfection. Total proteins were isolated with RPI solution. The extraction mixture was centrifuged at 12000 g at 4 °C for 20 min. The supernatants were transferred to a clean tube and stored at -70 °C as aliquots. The protein concentration was determined using Bradford dye-binding assay with bovine serum albumin as a standard. Total proteins were separated on 120 g/L SDS-polyacrylamide gel, and then transferred to nitrocellulose membranes. After being blocked for 1 h with the Tris/NaCl (50 mmol/L Tris-HCl, 200 mmol/L NaCl, 0.5 g/L Tween-20, pH7.4) containing 20 g/L BSA, the membranes were probed with the goat anti-mouse Prx I polyclonal antibody. Immunoreactive bands were visualized with solution containing 1 mg/mL DAB and 0.1 mL/L H2O2. The expression of Prx I was normalized by that of GAPDH.
RESULTS
Amplification of the cDNA of Prx I
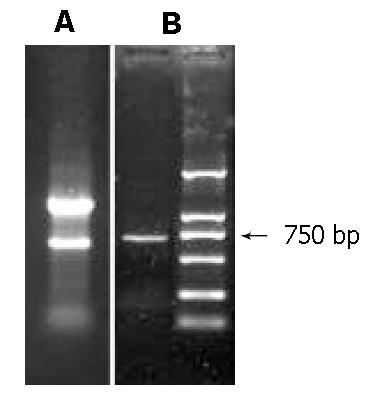
Total RNAs were isolated with 3 main bands appeared on agarose gels electrophoresis (Figure 1A). Message RNAs were smeared on the gel. After 28 cycles of RT-PCR, the products were run on 12 g/L agarose gel. A DNA band about 750 bp was visualized which was coincident with the length of the Prx I cDNA and could be cloned in the next step (Figure 1).
Figure 1 Amplification of Peroxiredoxin I by RT-PCR.
A: total RNAs were isolated from mouse intestinal epithelia; three bands were visualized on agarose gel, which represented the main ribosome RNA of the cells. B: amplification of the coding region of peroxiredoxin I applying RT-PCR.
Cloning of Prx I
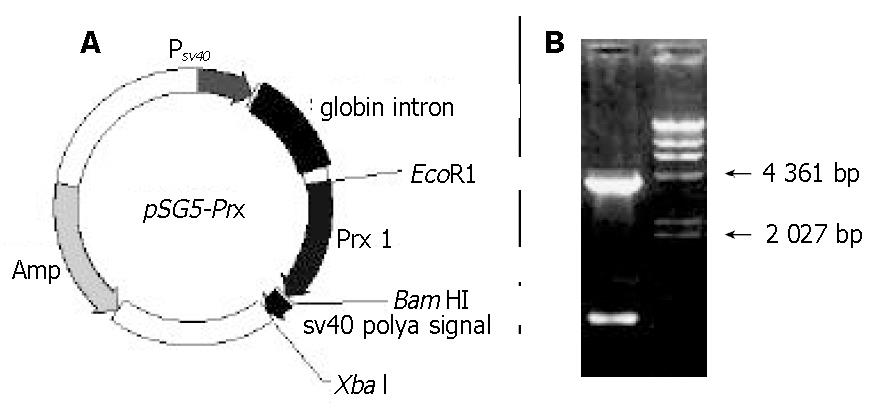
After the PCR product was ligated to T-vector, the linkage mixture transformed competent bacteria of JM109, and white colonies grew on LB plate containing ampicillin, X-gal and IPTG. Inserted DNA of 3 random colonies was sequenced, and the DNA was completely coincident with the sequence in GeneBank (data not shown). Then the inserted DNA of pT-Prx was digested with EcoR I and BamH I, purified, and inserted into vector pSG5. The recombinant expression vector was termed as pSG5-Prx (Figure 2). This recombinant plasmid was directly applied in cell transfection.
Figure 2 Recombinant expression vector pSG5-Prx.
A: diagrams of the expression vector pSG5-Prx. B: electrophoresis map of this vector (EcoR I and BamH I). λ/HindIII marker was loaded to estimate the relative molecular weight of the expression vector.
Transcription of pSG5-Prx in IEC-6 Cells by RT-PCR
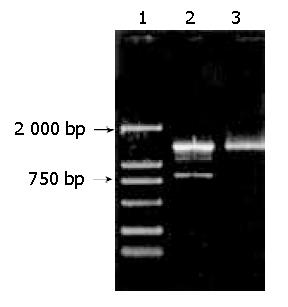
We transfected IEC-6 cells with pSG5-Prx using pSG5 as a control. The cells grew very well during the period of transfection. Total RNAs of transfected IEC-6 cells were isolated after the second transfection using TriPure solution. Transcription of the recombinant plasmid was detected by applying RT-PCR with primers P2 and P3. The result showed that 2 DNA bands were visualized on agarose gel. Because the beta-globin intron (Figure 2A) in the primary transcript was spliced, the length of RT-PCR product based on mature mRNA was about 817 bp. However, the length of the other DNA band was about 1399 bp, which was the PCR product using the recombinant plasmid as template (Figure 3).
Figure 3 Detection of the transcription of recombinant vector pSG5-Prx by RT-PCR.
Total RNAs isolated from transfected cells were applying for RT-PCR analysis with primers P2 and P3. Lane 1: DNA marker DL 2000, Lane 2: RT-PCR product, Lane 3: PCR product using purified recombinant vector as template.
Expression of Prx I in IEC-6 Cells
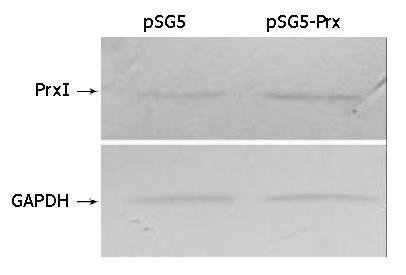
Total proteins of transfected IEC-6 cells were isolated after the second transfection and separated by SDS-PAGE. The blots were probed with specific antibodies against Prx I and GAPDH. Relative protein level was determined by quantitating the intensity of the bands with a densitometer. The ratio of the intensity of Prx I/GAPDH was automatically compared by Quantity One software (Bio-Rad Corp.). The result showed that the expression of Prx I was increased in pSG5-Prx transfected IEC-6 cells (50% more than the control cells) (Figure 4). This indicated that the recombinant plasmid could be transcripted and translated in IEC-6 cells.
Figure 4 Immunoblotting of peroxiredoxin I of transfected and normal IEC-6 cell.
DISCUSSION
IEC-6 cell line was derived from normal rat intestinal epithelia. Its living condition for stable passage was successfully established after trials. The cell line was testified to be the small intestinal epithelial cell by electron microscopy and immunohistochemistry, and has been applied to some related research work[22]. It is rational to apply this cell line in vitro to mimic the intestinal epithelia in vivo.
To further study the biochemical and physiological function of Prx I, we cloned the coding region of this protein and over expressed it in IEC-6 cells in this study. The cDNA of Prx I was first cloned in 1994 applying differential hybridization technique, which was named as MSP23[23]. Treatment with diethylmaleate or glucose/glucose oxidase markedly enhanced the induction of its transcription, and the mount of its protein in the cells rapidly increased. Moreover, the cDNAs corresponding to the thiol-specific antioxidant (TSA), which encoded a protein termed as Prx, has been cloned and sequenced in yeast and mammalian cells[24]. From the results of biochemical and physiological studies, Prx I has been involved in association with a variety of diverse cellular functions including proliferation, differentiation, and immune response. However, the detailed pathophysiological functions of this protein remain unknown.
The most striking finding of Prx I was that it was increased in many kinds of tumor cells. Moreover, some researchers thought it might be a potential tumor marker[25]. Prx I was over-expressed in human breast cancer tissues compared to normal tissues[26]. Using comparative proteome analysis between human normal (BEAS 2B) and malignant (A549) lung epithelial cells, Chang et al[27] found that Prx I was increased more than twofold reproducibly. Additionally, Prx I expression levels in follicular neoplasm and thyroiditis group were significantly higher than that of the control group, though papillary carcinoma group did not show statistical significance[25]. All these data suggested Prx I was involved in tumor development or progression, and it might be a new target for gene therapy of tumor cells.
It was suggested that Prx I was a radiation responsive gene. Lee et al[28] demonstrated, when the irradiated testis was examined, Prx I was found to be transiently up-regulated. Its highly homologous gene Prx II was increased in tissues isolated from the patients who did not respond to radiation therapy. However, treatment with a Prx II antisense decreased the expression of Prx II, enhancing the radiation sensitivity of cells[29].
In this study, we found pSG5 was an efficient expression vector of mammal cells[30]. Though it has no screen marker, the expression of recombinant plasmid can be easily verified at mRNA level applying RT-PCR technology owing to the globin intron of this vector. Because of the existence of the vector DNA in the sample purified by applying Tripure solution, two kinds of RT-PCR product were displayed on agarose gel, which represented different PCR template. After the second transfection, most of the cells expressed the exogenous vector, and the protein was accumulated in these cells, which could be detected by immunoblotting. We will further study the biochemical and physiological role in radiation response of this protein.