Published online Jul 15, 2004. doi: 10.3748/wjg.v10.i14.2050
Revised: February 26, 2004
Accepted: March 6, 2004
Published online: July 15, 2004
AIM: To investigate the role of human La protein in HBV mRNA expression.
METHODS: Three human La protein (hLa) specific siRNA expression cassettes (SECs) containing U6+1 promoter were prepared via one-step overlapping extension PCR. After transfection with SECs into HepG2 cells, inhibition effects on hLa expression were analyzed by semi-quantitative RT-PCR and Western blotting. Then, effective SECs were screened out and transfected into 2.2.15 cells, a stable HBV-producing cell line. HBV surface antigen (HBsAg) and e antigen (HBeAg) secretions into culture media were detected by microparticle enzyme immunoassay (MEIA) and HBs and HBe mRNA levels were analyzed by semi-quantitative RT-PCR.
RESULTS: SEC products containing U6+1 snRNA promoter, and 3 sites of hLa mRNA specific siRNA were obtained successfully by one-step overlapping extension PCR and could be directly transfected into HepG2 cells, resulting in inhibition of La protein expression in both mRNA and protein levels, among which U6+1-hLa833 was the most efficient, which reduced 18.6-fold mRNA and 89% protein level respectively. In 2.2.15 cells, U6+1-hLa833 was also efficient on inhibition of hLa expression. Furthermore, semi-quantitative RT-PCR showed that HBs and HBe mRNA levels were significantly decreased by 8- and 66-fold in U6+1-hLa833 transfected cells compared to control. Accordingly, HBsAg and HBeAg secretions were decreased partly posttransfection with SECs.
CONCLUSION: PCR-based SECs can be used to mediate RNAi in mammalian cells and provide a novel approach to study the function of La protein. The inhibition of La protein expression can result in a significant decrease of HBV mRNA, which implies that the hLa protein is also involved HBV RNA metabolism as one of the HBV RNA-stabilizing factors in human cells.
- Citation: Ni Q, Chen Z, Yao HP, Yang ZG, Liu KZ, Wu LL. Inhibition of human La protein by RNA interference downregulates hepatitis B virus mRNA in 2.2.15 cells. World J Gastroenterol 2004; 10(14): 2050-2054
- URL: https://www.wjgnet.com/1007-9327/full/v10/i14/2050.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i14.2050
Human La protein is a 47-ku phosphoprotein predominantly localized in nuclei. La protein is a member of RNA-binding proteins containing RNA recognition motifs (RRM) and interacts with RNA polymerase III transcripts such as pre-tRNA by binding to a small stretch of uridines at the 3’-end common to these transcripts and might be necessary for proper processing of these precursors[1-3]. In addition, La protein is known to interact with a variety of viral RNAs for stabilizing various RNAs[4-7], and is required for viral internal ribosomal entry site (IRES) -mediated translation[8-12].
La protein has been identified as a host factor potentially involved in the cytokine-induced post-transcriptional down-regulation of hepatitis B virus (HBV) RNA[13]. A strong correlation between cytokine-mediated disappearance of HBV RNA and cytokine-induced processing of full-length mouse La protein (mLa) was observed. The mLa binding site was mapped to a predicted stem-loop structure within a region located at the 5’-end of the post-transcriptional regulatory element of HBV shared by all HBV RNAs[5,6]. In addition, HBV RNA was accessible to endoribonucleolytic cleavage near this mLa binding site and HBV RNA substrates were more efficiently cleaved after induction of mLa processing[7]. All these findings indicate that La protein might be an HBV RNA-stabilizing factor. Determination of the high affinity interaction between human La protein (hLa) and HBV RNA in vitro[14], which is similar to that of mLa-HBV RNA interaction, implied that hLa might be involved in HBV RNA metabolism. But the role of human La protein (hLa) in HBV RNA metabolism in vivo is still unknown at present.
RNA interference (RNAi) is a process of sequence-specific post-transcriptional gene silencing via double-stranded RNA (dsRNA) present in plants and invertebrates[15]. With the increasing findings that 21-23 nt RNA duplexes known as small interfering RNA (siRNA) can also specifically and effectively knock down target gene expression in mammalian cells but short enough to evade host response[16], RNAi has been promptly developed into a powerful tool for studying protein function.
In this report, we utilized PCR-based siRNA strategy[17-19] to obtain several hLa-specific siRNA expression cassettes (SECs) containing U6+1 snRNA promoter and to deplete the hLa expression by transfection with SECs into human cells. In a stable HBV-producing cell line 2.2.15 cells[20], the inhibition of hLa expression resulted in a significant decrease of HBV mRNA and partly reduction of HBV antigen secretion. This result suggests that human La protein also plays an important role in HBV expression.
Pyrobest DNA polymerase was obtained from Takara Biotech (Japan). TaqPlus DNA polymerase was purchased from Dingguo (China). QIAquick PCR purification kit was obtained from Qiagen (Germany). M-MuLV reverse transcriptase was obtained from MBI fermentas (Lithuania). G418 was from Clontech (USA). Fetal calf serum was from Hyclone (USA). Trizol reagent and Lipofectamine 2000 were obtained from Invitrogen Lifetechnology (USA). Mouse monoclonal antibody against human La protein was purchased from BD Biosciences (USA). Rabbit polyclonal antibody against actin was from Wuhan Boster Biological Technology (China). Donkey anti-mouse and goat anti-rabbit horseradish peroxidase (HRP)-IgG were from SantaCruz (USA). Re-Blot Plus Western blot recycling kit was obtained from Chemicon (USA). SuperSignal west dura extended duration substrate ECL kit and CL-Xposure film were obtained from Pierce Biotech (USA). AxSYM and detective kits of HBsAg and HBeAg were purchased from ABBOTT (USA). Polyvinylidene difluoride (PVDF) membranes were obtained from Millipore (USA). All PCR primers were synthesized by Shanghai Sangon Biological Company (China).
Plasmid pAVU6+27 was a gift of Dr. Paul D. Good (Engelke Laboratory, Department of Biological Chemistry). Plasmid pcDNA3.1 was obtained from Clontech (USA). HepG2, a human hepatoblastoma cell line, and 2.2.15 cells derived from HepG2 cells and stably transfected with HBV DNA[20] were maintained in our laboratory.
Target sites of siRNA were determined by using on-line tool from Ambion Company. Three sites for hLa located downstream of the start codon were selected, their sequences are as follows: coding region 347-357 (5’-AAGGCTTCCCAACTGATGCAA-3’) for hLa347, 833-843 (5’-AAGCCAAGGAAGCATTGGGTA-3’) for hLa833, 911-921 (5’-AAGTACTAGAAGGAGAGGTGG-3’) for hLa911. 5’-AAGGGCGAGGAGCTGTTCACC-3’for EGFP10 siRNA site was used as control for studying the effect of hLa siRNA. Using 5’-TTCAAGAGA-3’ as 9-nt loop, all short hairpin RNAs (shRNA) were designed according to the structure of siRNA sense strand-loop-siRNA antisense strand.
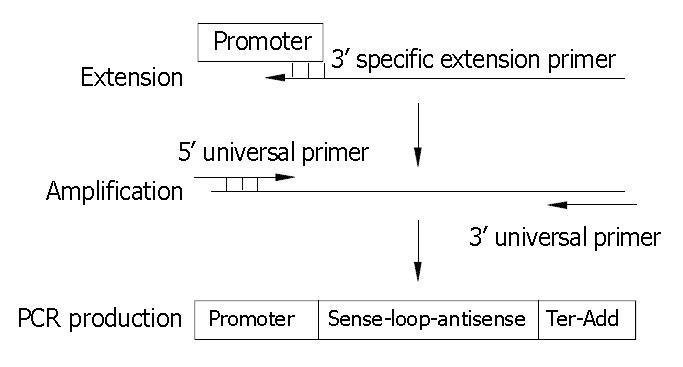
U6+1 promoter was obtained from plasmid pAVU6+27 containing human U6 promoter. Oligonucleotides coding hairpin shRNA antisense were synthesized and used for PCR reaction as 3’ specific extension primers. All extension primers have the 5’ end 21-nt containing additional adaptor sequence and a stretch of 6 deoxyadenosines as transcription terminator. The 5’ universal primer is complementary to 18 nt at the 5’ end of the U6 snRNA promoter (bold italics) 5’-GGAAGATCTGGATCCAAGGTCGGG-3’ and the 3’ universal primer is complementary to the first 21-nt at the 5’ end of all extension primers 5’-CGGCT CTAGAGTTCAAAAAAG-3’. Both universal primers were 5’-phosphorylated and could be used for all PCR reactions. SEC was constructed by using one specific extension primer and two universal primers via overlapping extension one-step PCR as depicted in Figure 1.
Using Pyrobest polymerase 30 cycles of PCR reactions were carried out, each at 94 °C for 30 s, at 58 °C for 30 s, and at 72 °C for 30 s. The PCR products were purified using QIAquick PCR purification kit (Qiagen) and stored at -20 °C prior to use.
HepG2 cells were maintained in Dulbecco’s modified Eagle’s minimal essential medium (DMEM) supplemented with 100 mL/L FCS, streptomycin (100 μg/mL) and penicillin (100 IU/mL) at 37 °C in a humidified atmosphere containing 50 mL/L CO2. 2.2.15 cells were grown in DMEM with 100 mL/L FCS, 400 μg/mL G418. HepG2 and 2.2.15 cells were seeded into 24-well plates 24 h or 48 h prior to transfection. HepG2 cells at 50% confluence and 2.2.15 cells at 70% were prepared for transfection. For all co-transfections, a total of 0.05 μg pcDNA3.1+ (used as carrier DNA) and 0.2 μg SEC was delivered using Lipofectamine 2000 according to the manufacturer’s instructions. HepG2 cells were harvested for analysis of La mRNA and protein expression. 2.2.15 cells and supernatants were harvested for detection of HBV mRNA and antigen.
Total RNA was extracted from cells with Trizol reagent according to the manufacturer’s instructions, 1.0 μg of total RNA was reverse transcribed to cDNA using oligo (dT) 18 as primer and Mu-MLV reverse transcriptase, and 0.5 μL of the cDNA template was separately used to amplify different mRNA. Information of primers is shown in Table 1. The PCR conditions for different target were as follows. A total of 28 cycles for GAPDH were performed, each at 94 °C for 30 s, at 60 °C for 30 s, and at 72 °C for 30 s. A total of 26 cycles for hLa were performed, each at 94 °C for 30 s, at 58 °C for 30 s, and at 72 °C for 30 s. A total of 26 cycles for HBs were performed, each at 94 °C for 30 s, at 56 °C for 30 s, and at 72 °C for 30 s. A total of 30 cycles for HBe were performed, each at 94 °C for 30 s, at 46 °C for 30 s, and at 72 °C for 45 s. After electrophoresis and scanning, all PCR product bands were analyzed by using the software Gel Pro analyzer32 and relative mRNA expression was estimated by normalization with GAPDH.
| Primer | GenBank accession No | Sequence of primer pair |
| GAPDH | BC023632 (26-260) | Sense: 5’-TGGGGAAGGTGAAGGTCGGA -3’ |
| Antisense: 5’- GGGATCTCGCTGCTCGAAGA-3’ | ||
| hLa | BC001289 (871-1313) | Sense: 5’-GGATAGACTTCGTCAGAGGAGCA -3’ |
| Antisense: 5’-CTGGTCTCCAGCACCATTTTCTG -3’ | ||
| HBs | U95551 (232-681) | Sense: 5’-CTCACAATACCGCAGAGTC-3’ |
| Antisense: 5’-TAAACTGAGCCAGGAGAAA-3’ | ||
| HBe | U95551 (1816-2452) | Sense: 5’-ATGCAACTTTTTCACCTC-3’ |
| Antisense: 5’-AACATTGAGGTTCCCGAG-3’ |
Cells were harvested and lysed in lysis buffer (0.1 mol/L Tris, pH 7.6, containing 0.15 mol/L NaCl, 2 mmol/L EDTA, 5 g/L Nonidet P-40, 5 g/L Triton X-100, 100 μmol/L sodium vanadate, 10 μg/mL aprotinin, and 20 μg/mL soybean trypsin inhibitor). The cell lysate (10-15 μg protein) was separated by sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel electrophoresis and electrophoretically transferred onto PVDF membrane. After non-specific binding sites were blocked with 50 g/L non-fat milk, the membrane was incubated with mouse anti-hLa monoclonal antibody (1:600 dilution) overnight at 4 °C. After washed, the blot was incubated with HRP-conjugated anti-mouse IgG for 1 h at room temperature, and immunoreactive bands were visualized with the ECL reagent. After the blot was stripped with stripping solution, the blot was reprobed with rabbit anti-actin polyclonal antibody for comparison of protein load in each lane. Densitometric scanning of the X-ray film following chemiluminescence was done and La protein expression level was estimated after normalization with actin.
Supernatants of 2.2.15 cells were harvested on days 1, 2, 3, 6 and 9 postransfection respectively in same wells. Each experiment well was performed in triplicate. A microparticle enzyme immunoassay (MEIA) method was applied for the detection of HBsAg and HBeAg according to the kit’s instructions. Sample with S/N values great than or equal to 2.0 for HBsAg, and S/CO values great than or equal to 2.1 for HBeAg were considered reactive as described previously[21].
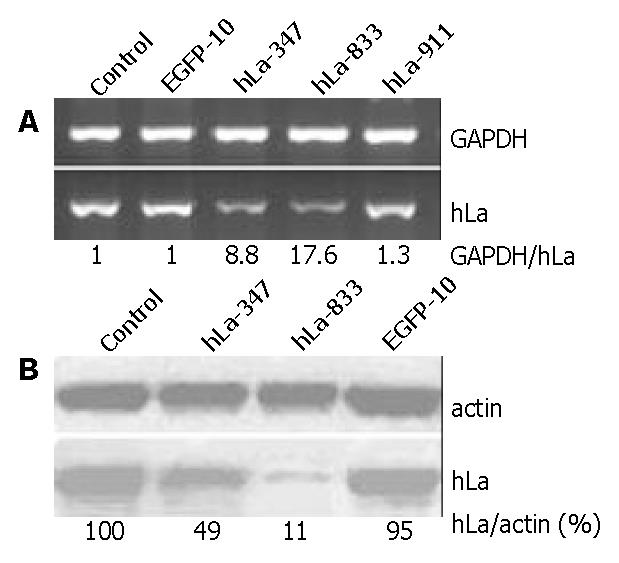
To determine whether siRNA specific to the hLa gene sequence could inhibit hLa expression, we screened the activity of three sites of hLa-specific siRNA by semi-quantitated RT-PCR of hLa mRNA in HepG2 cells 36 h posttransfection with SECs. Two of them, SEC U6+1-hLa347 and U6+1-hLa833 siginificantly decreased hLa mRNA levels by about 8.8- and 17.6-fold respectively as compared with the control (Figure 2A). However, SEC U6+1-hLa911 only slightly affected hLa mRNA expression (1.3-fold reduction). No significant inhibition of hLa mRNA expression was detected in cells transfected with SEC U6+1-EGFP10 (Figure 2A). Furthermore, we performed Western blot analysis to verify the inhibitory effects on hLa protein expression levels 72 h posttransfection with SEC U6+1-hLa347 and U6+1-hLa833. As shown in Figure 2B, hLa protein expression almost could not be detected in cells transfected with U6+1-hLa833. After normalized to actin, the level of hLa protein expression was reduced to approximately 11% and 49% of the control cells for U6+1-hLa833 and U6+1-hLa347 respectively, and no reduction was found for U6+1-EGFP10. These data demonstrated that SEC-mediated RNAi could effectively inhibit human La protein expression in cultured HepG2 cells and the down-regulation in hLa was sequence-specific and highly site-dependant.
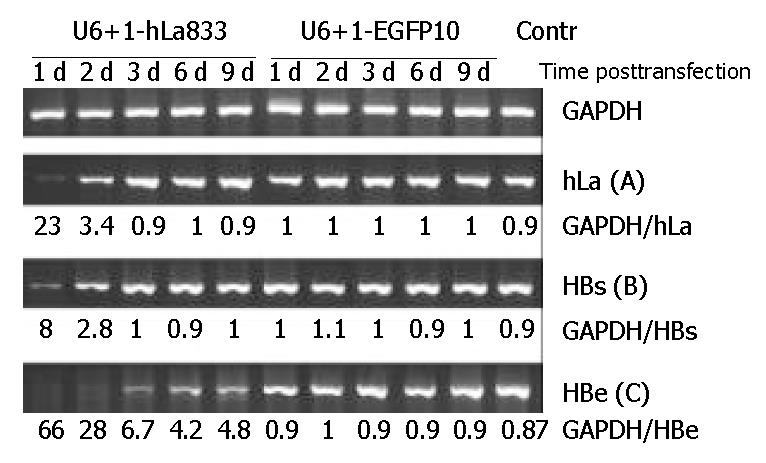
As SEC U6+1-hLa833 could greatly specific inhibit hLa mRNA and protein expressions in HepG2 cells, we then assessed its inhibitory effects on 2.2.15 cells derived from HepG2. As shown in Figure 3A, inhibition of hLa mRNA levels was detected with the reduction of about 22- and 3.4-fold on days 1 and 2 posttransfection, but no inhibitory effects were observed from days 3 to 9 posttransfection. In addition, no significant inhibition of hLa mRNA levels was detected in cells posttransfection with the SEC U6+1-EGFP-10 (Figure 3A).
This result indicates that SEC-mediated RNAi could also effectively inhibit hLa mRNA expression in cultured 2.2.15 cells, but the inhibitory effects could last only for a short time.
In order to investigate the relationship between hLa mRNA and HBV RNA expression, using the same samples, the levels of HBs and HBe mRNA expression in SEC-transfected 2.2.15 cells were detected by semi-quantitative RT-PCR. The HBs mRNA levels were also decreased about 8- and 2.8-fold in the first 2 d posttransfection, which were associated with a parallel reduction of hLa mRNA levels (Figure 3B). Moreover, HBe mRNA decreased dramatically, and almost could not be detected in the first 2 d and the inhibitory effect could last for at least 9 d (Figure 3C). In contrast, no significant changes in HBs and HBe mRNA levels were observed in cells posttransfection with SEC U6+1-EGFP-10 (Figure 3B and Figure 3C). These results demonstrated that RNAi against hLa could lead to reduction of HBV mRNA levels in 2.2.15 cells.
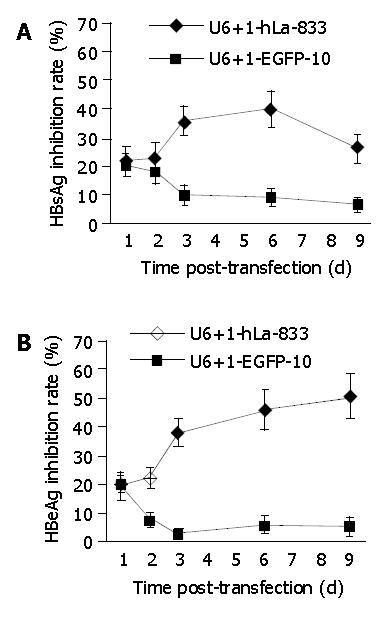
If the inhibition of hLa expression could affect HBs and HBe mRNA expressions in 2.2.15 cells, HBsAg and HBeAg secretion would be affected accordingly. Supernatant of SEC-transfected and control cells was harvested in the same period of posttransfection, HBsAg and HBeAg concentrations in the culture media were measured by MEIA method. Figure 4A shows that transfection with SEC U6+1-hLa833 inhibited the secretion of HBsAg in 2.2.15 cells from days 3 to 9 posttransfection in comparison with that of SEC U6+1-EGFP10 (P < 0.05). The maximal inhibitory effect on day 6 posttransfection was 40%, and returned to 26% on d 9. HBeAg secretion was inhibited 38% on d 3 posttransfection with SEC U6+1-hLa833 and remained 46% and 51% on d 6 and 9 respectively (Figure 4B). There was a significant difference (P < 0.05) in HBeAg secretion between SEC U6+1-hLa833- and U6+1-EGFP10-transfected cells on d 3, 6 and 9 posttransfection.
IR = [(χ of wells for control-χ of wells for thansfection)/(χ of wells for control - N)] × 100%
χ represents S/N or S/CO for HBsAg or HBeAg, and n = 2.0 for HBsAg and 2.1 for HBeAg respectively.
Results showed here from studies in 2.2.15 cells using siRNA against human La protein revealed a significant reduction in hLa expression, that resulted in HBV mRNA level reduction simultaneously, especially the expression of HBe mRNA, which was markedly decreased in the first 3 d and maintained at a lower level on the 9th d after inhibition of hLa expression. Following HBV mRNA reduction, HBsAg and HBeAg secretions were decreased accordingly though reduction of HBV antigen was less significant than that of corresponding mRNA. The relevance of RNAi-mediated knockdown of hLa towards reduction of HBV mRNA level suggested that human La protein might also be involved in HBV RNA metabolism. Based on the previous findings of the high affinity interaction between human La protein (hLa) and HBV RNA in vitro[14], and mouse La protein might be an HBV RNA-stabilizing factor in HBV-transgenic mouse model[5,7]. Our results invited a presumption that La protein might also play a role in stabilizing HBV RNA in human cells. However, HBsAg and HBeAg secretions could be affected slightly by hLa expression level, indicating that La protein is not a unique factor, and additional factors could contribute to viral RNA stability or HBV expression regulation. Therefore, using only hLa as targets for a novel antiviral strategy[14] would not be practical. Experiments to investigate the role of other RNA-binding proteins in HBV RNA metabolism might gain better insights into this problem.
We also observed that downregulation of HBe seemed more significant than HBs, implying more abundant HBs mRNA could generate HBsAg than HBe in 2.2.15 cells, which was in accord with the redundant HBsAg expression and secretion from patients with HBV infection. So HBs might be poorly susceptible to interfering factors. In addition, the feature of overlapping reading frames made entire 2.1-kb mRNA sequence encode HBs that was contained in 3.5-kb mRNA sequence encoding Hbe[22,23]. In fact, HBs mRNA level detected by RT-PCR in our experiments represented the total transcript level containing HBe and HBs. Therefore, instead of RT-PCR, Northern blot analysis should reflect the truth of all HBV transcripts in more detail.
As to siRNA application in RNAi technology, besides chemically synthesized 21-nt siRNA duplexes[16], some approaches could generate siRNA for various needs, such as in vitro transcribed siRNA[24,25], plasmid DNA expression vector-based siRNA[26,27], viral vector-based siRNA[28-30], and PCR-based siRNA expression cassette (SEC)[17-19] which was proved to be a rapid, facile and cheap approach for identification of optimal siRNA-targets. In this report, we attempted to simplify the commonly used two-step PCR into overlapping-extension one-step PCR reaction by using three primers: a long specific extension primer and two universal primers, and also successfully prepared SECs targeting three sites of hLa mRNA. After transfection with these SECs into HepG2 cells, SEC U6+1-hLa833 demonstrated that strong RNAi effect was promptly screened out as optimal SEC to be used for further study. As a 347-bp in length of SEC DNA, it is often difficult to transfect them into mammalian cells, but using plasmid as carrier DNA, the efficiency of transfection seemed to be improved significantly (data not shown). On the other hand, stability of SEC DNA in cells should be considered. Despite SECs were phosphorylated for enhancing resistance to nuclease in cells, but target mRNA level still returned to normal on day 3 posttransfection, which maintained RNAi effect not as long as reported in application of vector-based siRNA[26], indicating that SEC strategy should be just adapted to transient gene silencing at present and the better modification strategies should be developed for its stability.
To our knowledge, this is the first report that describes the role of human La protein in HBV expression in cultured human cells. Elucidating the mechanism of human La protein affecting HBV expression and replication will allow a deeper understanding of the interaction between host factors and HBV during HBV infection and clearance, and will provide useful clues for controlling HBV infection.
We thank Dr. Paul D. Good (Engelke Laboratory) for supplying plasmid pAVU6+27. We also thank Dr. Jian-Er Wo, Dr. Yu Chen and Dr. Jun-Bin Shao for technological support, and Dr. Edward Zumbika for English revision of this paper.
Edited by Wang XL Proofread by Chen WW and Xu FM
| 1. | Chakshusmathi G, Kim SD, Rubinson DA, Wolin SL. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003;22:6562-6572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 102] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 2. | Wolin SL, Cedervall T. The La protein. Annu Rev Biochem. 2002;71:375-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 336] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 3. | Maraia RJ. La protein and the trafficking of nascent RNA polymerase iii transcripts. J Cell Biol. 2001;153:F13-F18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 4. | Spångberg K, Wiklund L, Schwartz S. Binding of the La autoantigen to the hepatitis C virus 3' untranslated region protects the RNA from rapid degradation in vitro. J Gen Virol. 2001;82:113-120. [PubMed] |
| 5. | Heise T, Guidotti LG, Cavanaugh VJ, Chisari FV. Hepatitis B virus RNA-binding proteins associated with cytokine-induced clearance of viral RNA from the liver of transgenic mice. J Virol. 1999;73:474-481. [PubMed] |
| 6. | Heise T, Guidotti LG, Chisari FV. La autoantigen specifically recognizes a predicted stem-loop in hepatitis B virus RNA. J Virol. 1999;73:5767-5776. [PubMed] |
| 7. | Heise T, Guidotti LG, Chisari FV. Characterization of nuclear RNases that cleave hepatitis B virus RNA near the La protein binding site. J Virol. 2001;75:6874-6883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 8. | Pudi R, Abhiman S, Srinivasan N, Das S. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by specific interaction of independent regions of human La autoantigen. J Biol Chem. 2003;278:12231-12240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 66] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 9. | Ali N, Pruijn GJ, Kenan DJ, Keene JD, Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J Biol Chem. 2000;275:27531-27540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 126] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | Cheung P, Zhang M, Yuan J, Chau D, Yanagawa B, McManus B, Yang D. Specific interactions of HeLa cell proteins with Coxsackievirus B3 RNA: La autoantigen binds differentially to multiple sites within the 5' untranslated region. Virus Res. 2002;90:23-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 11. | Ray PS, Das S. La autoantigen is required for the internal ribosome entry site-mediated translation of Coxsackievirus B3 RNA. Nucleic Acids Res. 2002;30:4500-4508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | De Nova-Ocampo M, Villegas-Sepúlveda N, del Angel RM. Translation elongation factor-1alpha, La, and PTB interact with the 3' untranslated region of dengue 4 virus RNA. Virology. 2002;295:337-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 153] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 13. | Tsui LV, Guidotti LG, Ishikawa T, Chisari FV. Posttranscriptional clearance of hepatitis B virus RNA by cytotoxic T lymphocyte-activated hepatocytes. Proc Natl Acad Sci USA. 1995;92:12398-12402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 90] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 14. | Horke S, Reumann K, Rang A, Heise T. Molecular characterization of the human La protein.hepatitis B virus RNA.B interaction in vitro. J Biol Chem. 2002;277:34949-34958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Hannon GJ. RNA interference. Nature. 2002;418:244-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3058] [Cited by in RCA: 2898] [Article Influence: 126.0] [Reference Citation Analysis (0)] |
| 16. | Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6971] [Cited by in RCA: 7019] [Article Influence: 292.5] [Reference Citation Analysis (0)] |
| 17. | Castanotto D, Li H, Rossi JJ. Functional siRNA expression from transfected PCR products. RNA. 2002;8:1454-1460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 106] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 18. | Gou D, Jin N, Liu L. Gene silencing in mammalian cells by PCR-based short hairpin RNA. FEBS Lett. 2003;548:113-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Zheng L, Liu J, Batalov S, Zhou D, Orth A, Ding S, Schultz PG. An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc Natl Acad Sci USA. 2004;101:135-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 153] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 20. | Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 868] [Cited by in RCA: 939] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 21. | Chen Y, Wu W. Determination of low level HBsAg in serum by microparticle enzyme immunoassay. Hepatobiliary Pancreat Dis Int. 2002;1:262-264. [PubMed] |
| 22. | Moolla N, Kew M, Arbuthnot P. Regulatory elements of hepatitis B virus transcription. J Viral Hepat. 2002;9:323-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 165] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 23. | Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1094] [Cited by in RCA: 1113] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 24. | Anderson J, Banerjea A, Planelles V, Akkina R. Potent suppression of HIV type 1 infection by a short hairpin anti-CXCR4 siRNA. AIDS Res Hum Retroviruses. 2003;19:699-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 25. | Myers JW, Jones JT, Meyer T, Ferrell JE. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat Biotechnol. 2003;21:324-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 146] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 26. | Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat Biotechnol. 2002;20:505-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 578] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 27. | Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3486] [Cited by in RCA: 3462] [Article Influence: 150.5] [Reference Citation Analysis (0)] |
| 28. | Liu CM, Liu DP, Dong WJ, Liang CC. Retrovirus vector-mediated stable gene silencing in human cell. Biochem Biophys Res Commun. 2004;313:716-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 29. | Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1186] [Cited by in RCA: 1190] [Article Influence: 54.1] [Reference Citation Analysis (0)] |
| 30. | Zhao LJ, Jian H, Zhu H. Specific gene inhibition by adenovirus-mediated expression of small interfering RNA. Gene. 2003;316:137-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 58] [Article Influence: 2.8] [Reference Citation Analysis (0)] |