Published online Jul 15, 2004. doi: 10.3748/wjg.v10.i14.2029
Revised: December 15, 2003
Accepted: January 15, 2004
Published online: July 15, 2004
AIM: To express chimeric Fd (cFd) and chimeric light chain (cL) in E. coli respectively and refold them into chimeric Fab (cFab) antibody.
METHODS: cFd and cL genes were respectively inserted into the prokaryotic expression vector pET32a to construct recombinant vectors pET32a/cFd and pET32a/cL. Then, the competent E. coli cells were transformed by the recombinant vectors and induced by IPTG. Moreover, a large quantity of cFd and cL expression products were prepared and mixed with equal molar to refold into cFab by gradient dialysis. The refolded products were identified and analyzed by sodium SDS-PAGE, Western blotting, ELISA and HPLC.
RESULTS: High efficient prokaryotic expressions of both cFd and cL in the form of non-fusion protein were obtained with the expression levels of 28.3% and 32.3% of total bacteria proteins, respectively. Their relative molecular masses were all 24 ku or so, and both of them mainly existed in the form of inclusion bodies. In addition, cFd and cL were successfully refolded into cFab by gradient dialysis, with about 59.45% of recovery when the starting total protein concentration was 100 μg/mL. The renatured cFab could specifically bind to related antigen with high affinity.
CONCLUSION: The cFab antibody against human hepatoma was highly and efficiently expressed and refolded, which laid a solid foundation for studying its application in the treatment of hepatoma.
- Citation: Xing JL, Yang XM, Yao XY, Song F, Chen ZN. Prokaryotic expression and renaturation of engineering chimeric Fab antibody against human hepatoma. World J Gastroenterol 2004; 10(14): 2029-2033
- URL: https://www.wjgnet.com/1007-9327/full/v10/i14/2029.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i14.2029
The advent of monoclonal antibody (mAb) greatly promotes application of antibodies in various fields[1]. However, due to immunogenicity and comparatively high molecular mass, mouse derived complete antibody was more or less limited in the application of disease diagnosis and treatment[2]. The cFab antibody is about 50 ku, only 1/3 of full-length IgG. Because of good penetrating ability, better characteristics of pharmaceutical kinetics and good antigen-binding activity, cFab antibody has been used more and more widely[3-5]. Compared with complete antibody, the cFab has no Fc fragment so that non-specific binding is decreased greatly. In addition, the cFab could not produce antibody-dependent cellular cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC), so it was mostly used as the carrier for targeted delivery of drugs[6]. It could also be further reconstructed into engineering cF(ab)2 antibody[7]. In this paper, based on cloned VH and VL genes of mAb HAb18 against human hepatoma[8], we expressed cFd and cL in E. coli in the form of non-fusion protein and refolded them into cFab antibody. It was expected that a practical protocol could be established for the preparation of a large quantity of cFab against human hepatoma, which would lay a solid foundation for further studies of its application in hepatoma treatment.
The pComb3/cFab vector containing cFd and cL genes of mAb HAb18 was previously constructed in our laboratory. Prokaryotic expression vector pET32a (+) and competent E. coli JM109(DE3) were purchased from Novagen Inc (USA). T vector, PCR reagents, restriction endonucleases and T4 DNA ligase were from Takara Inc. (DaLian, China). The mAb HAb18, chimeric IgG antibody chHAb18 and HRP-HAb18 were previously prepared in our laboratory. IPTG, FITC-labeled and HRP-labeled goat anti-human IgG were from SABC Inc. (Luo Yang, China). Protein G affinity column was purchased from Pharmacia Inc (USA). Hepatoma cell line HHCC was from ATCC (Shanghai, China).
According to gene sequences of HAb18 cFd and cL which were previously constructed in our laboratory, PCR primers were designed by computer software Primer Premier 5.0, and relative restriction endonuclease sites were introduced into primers for the construction of prokaryotic non-fusion expression vectors of cFd and cL. All PCR primers were synthesized by Shenggong Inc. (Shanghai, China), and their sequences were as follows: cFd back: 5’GCGGAATTCATATGGTTAAGCTTGAAGAGTCTGGAGGAGGCTT 3’; cFd forward: 5’GGGGTCGACTCATTAACTAGTTTTGTCACAAGATTT GGGCT3’; cL back: 5’GCGGAATTCATATGAGTATTGTGA TGACCCAGACTCCCA3’; cL forward: 5’GGGCCTCGAGTCATTAACATTCACCTCTGTTGAAGCTCT3’. Underlined sequences are restriction sites Nde I, Sal I and Xho I.
With the vector pComb3/cFab as template, cFd and cL genes were amplified using related primers. The PCR products were purified by gel extraction. Then, the plasmid pET32a (+) and PCR amplified cFd or cL gene were digested by a pair of restriction endonucleases Nde I and Sal I or Nde I and Xho I. After the corresponding target fragments were purified by gel extraction, ligation, transformation and screening of the positive clones containing the recombinant vector pET32a/cFd or pET32a/cL were sequentially conducted. Finally, recombinant vectors were identified by restriction endonucleases digestion and DNA sequencing was completed by Shenggong Inc. (Shanghai, China).
The E. coli cells containing pET32a/cFd or pET32a/cL or pET32a (+) were respectively inoculated with 1:100 into 5 mL LB medium containing ampicillin (100 µg/mL). When absorbance of A600 nm was up to 0.8 or so, 1 mL E. coli cultures were obtained for further assay. Then, IPTG was added to left cultures with 1 mmol/L of final concentration and another 10 h induction of expression (250 r/min, 37 °C) was conducted. After being treated by boiling and centrifugation, all E. coli samples after and before induction were loaded onto 120 g/L SDS-PAGE gel for further analysis. At the same time, Western blotting was done by HRP-labeled goat anti-human IgG (H + L). In addition, E. coli cells containing pET32a/cFd and pET32a/cL after induction were collected and treated by repeated freezing and thawing. Then, the location of expression products was investigated by SDS-PAGE.
A total of 500 mL cultures of E. coli containing pET32a/cFd or pET32a/cL were induced for the protein expression under the same condition as mentioned above. Inclusion body was isolated and solubilized by 8 mol/L urea[9]. The total protein concentration was determined by bicinchoninic acid (BCA) assay[10] and the expression percentages of cFd and cL were evaluated by SDS-PAGE. cFd and cL dissolved in urea were mixed with equal molar, and the total protein concentration was adjusted to 100 µg/mL by adding the lysis solution. Renaturation was conducted by gradient dialysis as described by Lee et al[11]. The renatured products were centrifuged, and the supernatant was collected, then the precipitation was dissolved in the lysis solution again. Protein recovery was determined by BCA assay. Single cFd and single cL were set as control at the same refolding condition when cFab was renatured. All samples above were detected by SDS-PAGE, Western blotting and HPLC.
After the total protein concentration was respectively adjusted to 200 µg/mL and 400 µg/mL, cFab antibody was renatured according to the method described above. The protein concentration of all samples was detected by BCA assay and protein recovery was evaluated. In addition, all samples were loaded onto SDS-PAGE gel for further analysis.
After dialysis in PBS overnight at 4 °C, renatured cFab was purified by protein G affinity chromatography according to manufacturer’s protocol.
Detection of antigen-binding activity of purified cFab antibody Indirect ELISA GST-HAb18GE, GST-fusion expression product of extracellular region of HAb18G[12] was the antigen of mAb HAb18, and the purified GST expression products were coated on the ELISA plates for detecting the antigen-binding activity of purified cFab antibody. Detailed protocol was described by Yan et al[13].
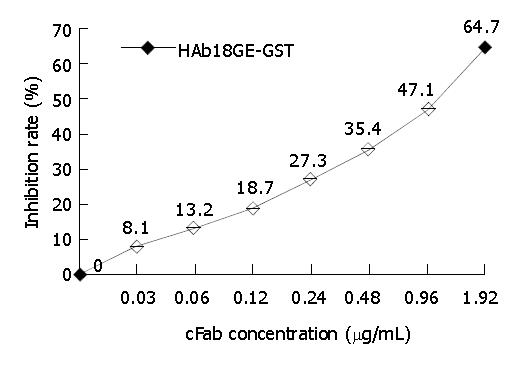
Competitive binding assay GST-HAb18GE was coated on ELISA plates overnight at 4 °C. After blocking, the mixture of HRP-HAb18 (0.1 mg/L) and gradiently diluted cFab was added into ELISA plates. After incubation for 1 h at 37 °C, the unbound antibody was washed away by PBST, followed by TMB staining. Finally, A450 was detected by an ELISA plate reader (Bio-Rad, USA), and inhibition rate and comparative affinity were determined according to the following method: Inhibition rate (%) = [(A450 of control group - A450 of experimental group)/A450 of control group] × 100%; comparative affinity (%) = [concentration of cFab when inhibition rate is 50%/concentration of HRP-HAb18(0.1 mg/L)] × 100%.
FACS detection The single-cell suspension of hepatoma cell line HHCC overexpressing HAb18G was prepared. The cell density was adjusted to 5 × 109 - 1 × 1010/L. HAb18 cFab diluted with horse serum was added into 50 μL HHCC single-cell suspension. Then, the mixture was incubated for 30 min at 4 °C. After washed twice by PBS, FITC-labeled rabbit anti-human IgG was added and the reaction mixture was incubated for 30 min at 4 °C. Then, washing and detecting were performed. In this assay, human IgG was set as negative control and PBS was set as blank control.
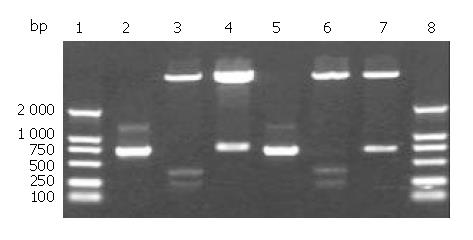
cFd and cL genes were successfully amplified using designed primers. Agarose gel electrophoresis showed that the sizes of cFd and cL genes were about 700 bp and 680 bp, respectively (lanes 2,5, Figure 1), which was in accordance with their expected sizes. The recombinant vectors pET32a/cFd and pET32a/cL digested by restrictive endonucleases (Figure 1) indicated that cFd and cL genes were correctly inserted into corresponding cloning sites. The results of DNA sequencing (results not provided) proved that cFd and cL genes inserted in expression vector had no base mutation and the codon reading frame was completely exact.
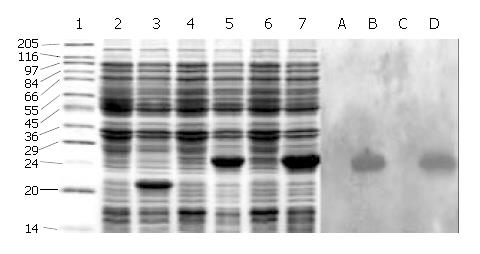
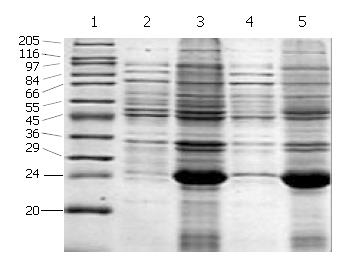
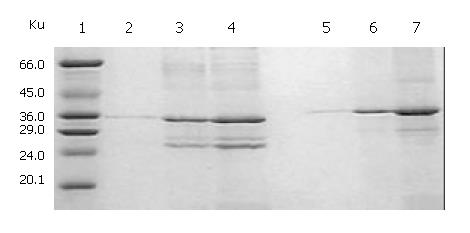
SDS-PAGE (Lanes 1-7, Figure 2) showed that a 21-ku new protein was expressed in E. coli transformed by control vector after induction, which is in accordance with the expression characteristics of the control vector, and that the recombinant cFd and cL with the same molecular weight of 24 ku were also successfully expressed after induced by IPTG. Scanning analysis by Smartview software indicated that the quantities of expressed cFd and cL were about 28.3% and 32.3% of the total bacterial protein, respectively. Western blotting (Lanes A-D, Figure 2) identified that both expressed cFd and cL were able to bind to anti-human IgG and the staining bands were at 24 ku. In addition, SDS-PAGE showed that the interested expression products existed in the form of insoluble inclusion body (Figure 3).
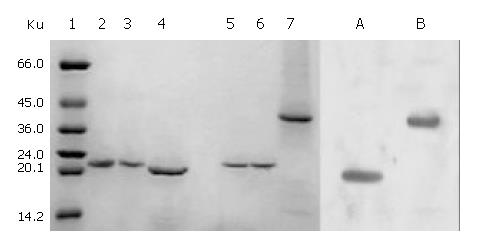
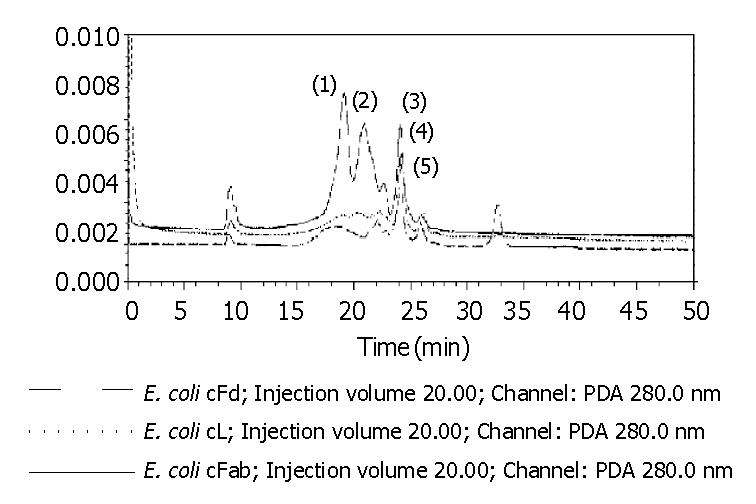
The total protein concentrations of inclusion body solution in 8 mol/L urea containing cFd or cL determined by BCA assay were respectively 12.04 mg/mL and 9.66 mg/mL. The results (not provided) of SDS-PAGE analysis indicated that cFd and cL were up to 49.8% and 58.3% in the total protein of inclusion body solution, respectively. SDS-PAGE (Figure 4, lanes 1-7) also showed that the new 45-ku protein band appeared under nonreduced condition after cFd and cL were mixed to refold. Western blotting (lanes A, B, Figure 4) also identified that the new protein bands could bind to anti-human IgG under reduced and nonreduced conditions. The recovery rate was about 75% when the starting concentration of total protein was 100 µg/mL. In addition, when the starting total protein concentration was separately increased to 200 µg/mL and 400 µg/mL, recovery rate was respectively decreased to 70.5% and 61%. SDS-PAGE (Figure 5) demonstrated that precipitation rose rapidly with the increase of starting total protein concentration, and the percentage of elevated precipitation was more than that of increased starting total protein concentration. HPLC analysis (Figure 6) showed that two new elution peaks (1) and (2) emerged after cFd and cL were mixed to refold in comparison with single refolded cFd and cL. The largest absorbance value of elution peak (2) was at 260 nm, so it was possible that elution peak (2) was not protein peak. In addition, the position of elution peak (3) was the same as elution peak (4) of refolded cFd and elution peak (5) of refolded cL. ELISA(result not provided) demonstrated that only elution peak (1) had specific antigen-binding activity. The peak areas of elution peak (1) and (3) were 33.49% and 17.75%, respectively. Renaturation yield of cFab antibody was about 49% determined by the method described previously.
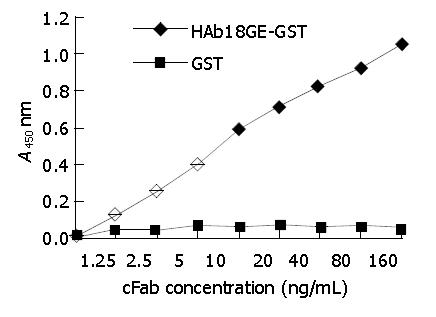
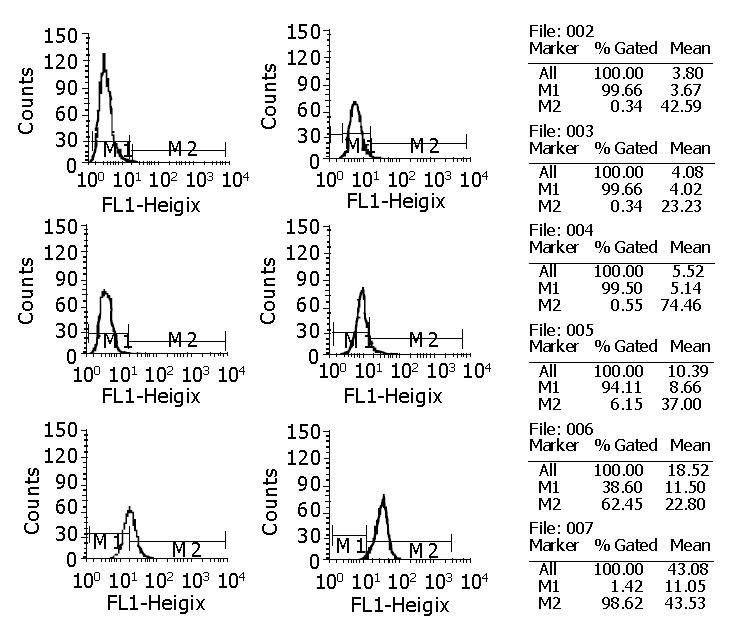
cFab 1 mL (80 µg/mL) was purified by protein G affinity chromatography. Indirect ELISA (Figure 7) showed that cFab was capable of specific binding to HAb18GE and the antigen-binding activity became higher with increasing concentration of cFab antibody. Competitive ELISA (Figure 8) indicated that the refolded cFab could competitively bind to the same antigen epitope as parental mouse antibody HAb18 and the binding ability improved with the increase of cFab antibody concentration. The affinity of cFab was evaluated to be 10% of parental mouse antibody HAb18 according to the concentration ratio of cFab and HAb18 when inhibition rate was 50%. FACS detection (Figure 9) demonstrated that cFab could bind specifically to hepatoma cell line HHCC and the binding capacity increased with the elevation of cFab antibody concentration.
In a previous study, we successfully expressed cFab antibody gene in the periplasmic compartment of E. coli. However, the productivity was rather low. So, we tried to improve the expression level of cFab. We separately expressed cFd and cL without signal peptide in cytoplasm of E. coli in the form of inclusion body. The expression levels of different fragment of antibody genes in E. coli were greatly diverse, which mainly depended on the design of translation initiation site, the type of antibody fragment, antibody gene sequence, sensitivity to proteases of expression products and the class of host bacterium, etc[14]. But high efficient expression of antibody genes was relatively easy to be obtained using prokaryotic expression vector. We successfully constructed recombinant vectors pET32a/cFd and pET32a/cL by cloning cFd and cL genes into the pET32a expression vector, and achieved high expression level of cFd and cL in E. coli. The expression products mainly existed in the form of the inclusion body, with 28.3% and 32.3% of expression percentages of total bacterial protein, respectively. Expression proteins in inclusion body could avoid being digested by proteases and were easy to be isolated and purified. cFd and cL must be refolded together in order to get active cFab.
As we know, effectively promoting the correct formation of intramolecular or intermolecular disulfide bond and inhibiting the formation of aggregation were key to protein renaturation[15]. In order to promote the refolded protein to form the correct disulfide bond, some reducers, such as β-sulfhydryl ethanol and DTT, were added into detergent solution to unfold wrong disulfide bonds of expression proteins in inclusion body. These wrong disulfide bonds might be formed during the lysis of host bacterium or during the resolving process of expression protein. When the protein was renatured, correct disulfide bonds could be reformed only by air oxidation[16]. In most cases, iron can promote the process of air oxidation[17]. In addition, the repeated formation and unclosing of the disulfide bonds may increase the probability of obtaining protein with correct conformation. This process could be facilitated by some enzymes (such as disulfide isomerase)[18] or oxidation-reduction response caused by the oxidation- and reduction-type sulfhydryl mixture[17] (such as glutathione). During the renaturation, the formation of aggregation was the most common reason resulting in lower renaturation efficiency. However, the mechanism of aggregation formation is still unclear. De Bern ardez Clark et al[19] suggested that the hydrophobic action among protein molecules was the most likely explanation for the aggregation formation facilitated by the formation of wrong disulfide bonds. In the experiment of mixed protein renaturation involving two or more proteins, small aggregation was first caused by the hydrophobic action, and then by the disulfide bonds formation from unpaired cystines that resulted in the formation of big heterogeneous aggregation. The formation of large quantities of aggregation would result in more protein precipitation, therefore, renaturation efficiency decreased greatly[11]. The previous study on renaturation also showed that arginine, a kind of stable reagent promoting protein dissolving during renaturation, could facilitate the correct folding of proteins possibly by inhibiting the formation of bigger aggregation[20].
In this study, we mixed cFd and cL with equal molar. The detergent was slowly removed from the mixture by gradient dialysis in the presence of a given concentration of arginine, while glutathione was added into the mixture to delete wrong reduced disulfide bonds. The results showed that this strategy for renaturation was successful. At the starting total protein concentration of 100 µg/mL, cFd and cL were refolded into cFab with 75% of protein recovery rate. Protein loss was mainly due to protein precipitation and adsorption onto dialysis belt. Replacing dialysis materials could reduce the latter loss. We identified the renatured cFab by SDS-PAGE, Western blotting and ELISA. The results proved that cFd and cL were successfully refolded into cFab. SDS-PAGE indicated that the molecular weight of renatured cFab was about 45 ku under the unreduced condition, less than 48 ku (cFd + cL), which revealed that the formation of intramolecular disulfide bond happened in the renatured protein[15]. HPLC of renatured products revealed that the renatured cFab produced two very different elution peaks. ELISA demonstrated that only elution peak (1) had antigen-binding activity. In addition, the biggest absorbance value of elution peak (2) was at 260 nm, which revealed that elution peak (2) might be nuclear acid existing in inclusion body. It was estimated by HPLC that the renaturation efficiency was up to 49%, which was quite desirable in comparison with the generally 10%-40% of renaturation efficiency for antibody protein. The desirable renaturation efficiency was mainly explained by good renaturation strategy and special antibody sequences, which are in accordance with the viewpoint of Wibbenmeyer et al[21]. It was also estimated that dozens of milligrams of active cFab could be gained from 1 L of E. coli cultures under the experimental condition described above. Therefore, further optimizing the preparation protocol can increase the yield.
When preparing a large quantity of active protein by dialysis renaturation method, another important influence factor must be taken into consideration, namely the volume of renaturation protein solution. The speed of protein aggregating and precipitating was positively correlated with the protein concentration, so protein precipitation would be predominant course when protein concentration was up to the highest point. As a result, the starting total protein concentration was generally very low, which would inevitably result in excessively large sample volume to be dealt with otherwise more troubles would arise. In this study, in order to explore the optimal renaturation condition of preparing a large quantity of cFab antibody, we increased the starting total protein concentration for renaturation to 200 µg/mL and 400 µg/mL, respectively, and investigated the renaturation efficiency after the protein concentration was increased. The results revealed that protein precipitation promptly increased with the increase of starting protein concentration, and increased percentage was more than that of protein concentration increase, which indicated that a part of renaturation efficiency was surely sacrificed in order to reduce the treatment volume and increase the speed of preparation during the large-scale production of cFab.
In conclusion, we successfully obtained cFab antibody with good activity by prokaryotic expression technique, followed by dialysis, which laid a solid foundation for further application in hepatoma treatment. In addition, based on this achievement, we are planning to modify cFab genes to produce another kind of more useful engineering antibody, bivalent chimeric F (ab’) 2[22]. It is expected to replace murine-derived F (ab’) 2 antibody produced by protease digestion, and reduce clinical cost and improve therapeutic efficacy.
Edited by Chen WW Proofread by Zhu LH and Xu FM
| 1. | Chen ZN, Bian HJ, Jiang JL. Recent progress in anti-hepatoma monoclonal antibody and its application. Huaren Xiaohua Zazhi. 1998;6:461-462. |
| 2. | Hasholzner U, Stieber P, Meier W, Lamerz R. Value of HAMA--determination in clinical practice--an overview. Anticancer Res. 1997;17:3055-3058. [PubMed] |
| 3. | Quinn MJ, Plow EF, Topol EJ. Platelet glycoprotein IIb/IIIa inhibitors: recognition of a two-edged sword. Circulation. 2002;106:379-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Lai ZZ, Xiong DS, Fan DM, Peng H, Xu YF, Zhu ZP, Yang YZ. High expression of the chimeric antibody Fab against-CD20 in E coli and bioactivity determination. Zhongguo Mianyixue Zazhi. 2000;16:521-524. |
| 5. | Xia L, Gu J, Zhang X, Liu Y, Wan H, Li P, Ruan C. Preparation of an antifibrin thrombus-specific murine/human chimeric monoclonal antibody Fab fragment in Escherichia coli. Thromb Res. 1996;81:477-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 6. | Better M, Bernhard SL, Lei SP, Fishwild DM, Lane JA, Carroll SF, Horwitz AH. Potent anti-CD5 ricin A chain immunoconjugates from bacterially produced Fab' and F(ab')2. Proc Natl Acad Sci U S A. 1993;90:457-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 25] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 7. | Humphreys DP, Vetterlein OM, Chapman AP, King DJ, Antoniw P, Suitters AJ, Reeks DG, Parton TA, King LM, Smith BJ. F(ab')2 molecules made from Escherichia coli produced Fab' with hinge sequences conferring increased serum survival in an animal model. J Immunol Methods. 1998;217:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Lou C, Chen ZN, Bian HJ, Li J, Zhou SB. Pharmacokinetics of radioimmunotherapeutic agent of direct labeling mAb 188Re-HAb18. World J Gastroenterol. 2002;8:69-73. [PubMed] |
| 9. | Wu JY, Jiang YH, Wang J, Zhang HB, Li M. Purification of recombinant human angiogenin. Guangdong Yixue. 1999;20:85. |
| 10. | Guo YJ, Wu D, Chen RW, Sun SH. [Cloning, high level expression and purification of porcine IFN gamma]. Shengwu Gongcheng Xuebao. 2001;17:183-186. [PubMed] |
| 11. | Lee MH, Kwak JW. Expression and functional reconstitution of a recombinant antibody (Fab') specific for human apolipoprotein B-100. J Biotechnol. 2003;101:189-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 12. | Xing JL, Chen ZN, Mi L, Li Y, Feng Q. Establishment of CHO cell strain that can highly efficiently express hepatoma associ-ate antigen HAb18G. Zhongliu. 2001;21:4-7. |
| 13. | Yan ZY, Wang HL. Jingbian Fenzishengwuxue Shiyan Zhinan [M]. Beijing: Science Press. 1998;. |
| 14. | Laden JC, Philibert P, Torreilles F, Pugnière M, Martineau P. Expression and folding of an antibody fragment selected in vivo for high expression levels in Escherichia coli cytoplasm. Res Microbiol. 2002;153:469-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 15. | Middelberg AP. Preparative protein refolding. Trends Biotechnol. 2002;20:437-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 260] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 16. | ANFINSEN CB, HABER E, SELA M, WHITE FH. The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc Natl Acad Sci U S A. 1961;47:1309-1314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 818] [Cited by in RCA: 677] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 17. | Saxena VP, Wetlaufer DB. Formation of three-dimensional structure in proteins. I. Rapid nonenzymic reactivation of reduced lysozyme. Biochemistry. 1970;9:5015-5023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 311] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 18. | Carmichael DF, Morin JE, Dixon JE. Purification and characterization of a thiol: protein disulfide oxidoreductase from bovine liver. J Biol Chem. 1977;252:7163-7167. [PubMed] |
| 19. | De Bernardez Clark E, Hevehan D, Szela S, Maachupalli-Reddy J. Oxidative renaturation of hen egg-white lysozyme. Folding vs aggregation. Biotechnol Prog. 1998;14:47-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 92] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 20. | Timasheff SN, Arakawa T. Stabilization of protein structure by solvents. In: Creighton TE. Ed. Protein Structure: A Practi-cal Approach, 2nd ed. Oxford: IRL Press. 1997;. |
| 21. | Wibbenmeyer JA, Xavier KA, Smith-Gill SJ, Willson RC. Cloning, expression, and characterization of the Fab fragment of the anti-lysozyme antibody HyHEL-5. Biochim Biophys Acta. 1999;1430:191-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |