Published online May 15, 2004. doi: 10.3748/wjg.v10.i10.1528
Revised: January 10, 2004
Accepted: February 1, 2004
Published online: May 15, 2004
AIM: To study the immune response of host to Helicobacter pylori VacA.
METHODS: The monocyte/macrophage-like U937 cells were infected with Helicobacter pylori vacA-positive strain NCTC 11638 or isogenic vacA-negative mutant. Differentially expressed genes were identified at 2, 6, 10, and 24 h post-infection by cDNA microarray. Differential expressions of some genes were confirmed by Northern blot.
RESULTS: More than 100 genes altered their mRNA expression at different time points respectively, many of which were identified to be related to immune evasion.
CONCLUSION: VacA is a crucial element for H pylori to escape from host immune defense by means of differentially regulating the expression of some related genes. These genes, previously known or unknown to be involved in the mechanism of immune evasion, deserve further investigation to unearth much more information complicated in the immune response.
-
Citation: Yuan JP, Li T, Li ZH, Yang GZ, Hu BY, Shi XD, Shi TL, Tong SQ, Guo XK. mRNA expression profiling reveals a role of
Helicobacter pylori vacuolating toxin in escaping host defense. World J Gastroenterol 2004; 10(10): 1528-1532 - URL: https://www.wjgnet.com/1007-9327/full/v10/i10/1528.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i10.1528
Helicobacter pylori infects about half of the world’s population. Despite the induction of an immunological reaction, the infection of H pylori is commonly life-long, suggesting that this pathogen has evolved mechanisms to evade protective immune responses to achieve the state of host-microbial equilibrium[1]. Some products of H pylori have been determined to have immunosuppressive effects for prolonging the infection. A 100 ku H pylori protein inhibits proliferation of T-cell and macrophage[2], and VacA, a cytotoxin that has been found to cause massive vacuolation in several mammalian cell lines and in the gastric epithelia of patients with active chronic gastritis associated with H pylori infection[3], may perform targeted action to disable T cells. VacA interferes with proteolytic processing of tetanus toxin and toxoid and specifically inhibits the Ii-dependent pathway of antigen presentation mediated by newly synthesized major histocompatibility complex (MHC) class II, suggesting that VacA may contribute to the persistence of H pylori by interfering with protective immunity[4]. However, to the author’s knowledge, the exact mechanism of such an immunosuppression effect has not been fully studied. Hence, in this study, we performed a large scale measurement of gene expression alteration in host cells using gene microarray technology, which provided the crucial information for interpreting the mechanisms of immunosuppression.
H pylori NCTC 11638 strain positive for vacA was given as a gift by Dr. Shi (Shanghai Institute of Gastroenterology). Isogenic vacA-negative mutant 11638-△ vacA was constructed by substitution of a kanamycin resistant gene for a short fragment of vacA through homologous recombination, as described previously[5]. H pylori strains were cultured routinely on brain heart infusion (BHI) agar plates with 5% sheep blood in mixed air containing 100 ml/L CO2, 50 mL/L O2, and 850 mL/L N2 at 37 °C.
The U937 cells were maintained in RPMI 1640 medium (Gibco BRL, USA) with 2 mmol/L L-glutamine, 1.5 g/L sodium bicarbonate, 4.5 g/L glucose, 10 mmol/L HEPES, 1.0 mmol/L sodium pyruvate and 100 ml/L fetal bovine serum. The day before H pylori infection, fresh medium with 20 mL/L fetal calf serum was substituted. Eighteen hours later, the cells grown to 90% confluency were cocultured with H pylori isogenic strains at a multiplicity of infection of 10 in culture medium for 2, 6, 10, and 24 h.
U937 cells cocultured with NCTC 11638 and 11638-△ vacA were collected at 2, 6, 10, and 24 h after the infection for mRNA extraction. Total RNA was isolated using TRIZOL reagent (Invitrogen, USA) according to the manufacturer’s instruction.
The cDNA microarrays were designed by Shanghai BioStar Genechip Inc. In this study, microarrays with 8464 human cDNAs were used, including full-length and partial complementary DNAs representing novel, known, and control genes.
Aliquots of 30 μg of total RNA were fluorescently labeled with Cy5- or Cy3-dCTP (Amersham Pharmacia, Sweden) by reverse transcription in the presence of 5 μg of oligo (dT) and 1 μL of SuperScriptII (Gibco-BRL, USA). The labeled cDNAs were purified using MicroSpin S-200 columns (Amersham Pharmacia, Sweden) and lyophilized. The probes were resuspended in 20 μL hybridization solution containing 8 μg of poly (dA), 2 μg of yeast tRNA, 10 μg of human Cot I DNA (Gibco-BRL, USA). After heated to 95 °C for 2 min and then cooled to room temperature, the mixture was applied to the slides and covered by a coverslip. The slides were incubated in a humid cabinet of an incubator for 16-18 h at 42 °C. Then the slides were washed at 60 °C for 10 min in solutions of 2 × SSC with 2 g/L SDS, 0.1 × SSC with 2 g/L SDS, and 0.1 × SSC sequentially, and then dried at room temperature.
Each slide was scanned at 10 μm resolution on a GenePix 4000B scanner (Axon Instruments, Inc., Foster City, CA) at variable PMT voltage to obtain maximal signal intensities with no more than 1% probe saturation. The images were processed with GenePix Pro 3.0. Ratios were normalized by a linear regression between ln(Cy5) and ln(Cy3) of all the genes’ background-corrected signal intensities on the microarray. Genes exhibited a 2-fold or greater change in expression level and exceeded 200 in signal intensity were considered true outliers.
The plasmids containing cDNA clone used for preparing probes were provided by Shanghai BioStar Genechip Inc. A total amount of 200 ng plasmids were used as templates for PCR amplification. PCR products were purified using QIAquick Gel Extraction Kit (Qiagen, Germany). The probes were labeled using random primed Strip-EZ DNA Kit (Ambion, Austin, TX). A total amount of 25 ng purified DNA diluted in 9 μL TE (10 mmol/L Tris-HCl, pH8, 1 mmol/L EDTA) was denatured at 95 °C for 5 min and then immediately frozen in liquid nitrogen, thawed, microfuged, and placed on ice. Afterwards, the following reaction was assembled as follows and mixed gently: A 9.0 μL of denatured DNA, 2.5 μL of 10 × Decamer solution, 5.0 μL of 5 × buffer-dATP/-dCTP, 2.5 μL of 10 × dCTP, 5.0 μL of [α-32P]dATP (3000 Ci/mmol, 10 mCi/mL), 1.0 μL of exonuclease-free Klenow, and nuclease-free water to 25 μL. After 20 min incubation at 37 °C, 1 μL of 0.5 mol/L EDTA was added to stop the reaction.
A 10 μg sample of total RNA per lane was subjected to electrophoresis on 12 g/L agarose gels containing 2.2 mol/L formaldehyde. RNAs were transferred onto Zeta-probe blotting membranes (Bio-Rad Laboratories, Hercules, CA) using Vacuum Blotter (model 785, Bio-Rad Laboratories) and baked under vacuum at 80 °C for 2 h. Membranes were hybridized for 16 h at 60 °C with ULTRArray hybridization solution (Ambion, Austin, TX) containing cDNA probes labeled with [α-32P] dCTP by random priming (Strip-EZ DNA labeling system, Ambion). The hybridized membranes were serially washed at 55 °C using 2 × SSC with 1 g/L SDS solution, then exposed to a phosphoimager. Blots were scanned and quantified by a phosphoimager in combination with Optiquant software v. 2.50 (Cyclone Storage Phosphor System, Packard Instruments).
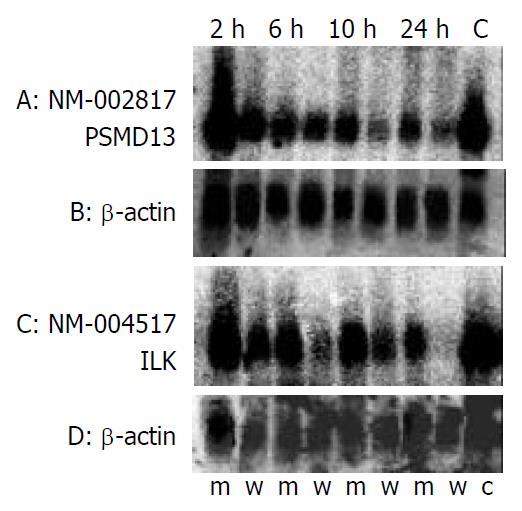
The mRNA expression in U937 cells was compared at 2, 6, 10, and 24 h after the infection with NCTC 11638 or 11638-△ vacA. More than 100 genes altered their mRNA expression at different time points respectively, among which, the genes related to immune evasion were selected (Table 1). To confirm the differential expression profiling of the cDNA microarrays, two genes: PSMD13 and ILK were chosen for northern blot analysis. As shown in Figure 1, the expression levels of these 2 genes were lower in the cells treated with H pylori NCTC11638 than those treated with 11638-△ vacA. In addition, in both cases, the fold ratio changes detected by the microarray were confirmed by Northern blot (Table 2). The level of β-actin transcript was present at approximately the same level in both samples, providing an assessment of RNA content in each sample used for Northern blot analysis.
| GeneBank_ID | Definition | Fold change | |||
| 2 h | 6 h | 10 h | 24 h | ||
| NM_004517 | Integrin-linked kinase (ILK) | 0.717 | 0.218 | 0.245 | 0.454 |
| NM_001964 | Early growth response 1 (EGR1) | 3.184 | 1.149 | — | — - |
| NM_022555 | Major histocompatibility complex, class II, DR beta 3 (HLA-DRB3) | 1.000 | 0.448 | 0.518 | 0.621 |
| NM_002116 | Major histocompatibility complex, class I, A (HLA-A) | 0.584 | 0.322 | 0.336 | — - |
| NM_002124 | Major histocompatibility complex, class II, DR beta 1 (HLA-DRB1) | — | 0.661 | 0.485 | 0.567 |
| NM_002817 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 (PSMD13) | 0.445 | 0.122 | 0.333 | 0.402 |
| NM_004159 | Proteasome (prosome, macropain) subunit, beta type, 8 | 0.573 | 0.150 | 0.304 | 0.254 |
| (large multifunctional protease 7) (PSMB8) | |||||
| NM_000616 | CD4 antigen (p55) (CD4) | 0.855 | 0.560 | 0.317 | 0.831 |
| NM_015953 | eNOS interacting protein (NOSIP), | 0.857 | 0.517 | 0.450 | 0.463 |
| NM_020998 | Macrophage stimulating 1 (hepatocyte growth factor-like) (MST1) | 0.690 | 0.562 | 0.353 | — - |
| NM_005944 | Antigen identified by monoclonal antibody MRC OX-2 (MOX2) | 1.514 | 1.027 | 2.042 | — - |
| NM_021138 | TNF receptor-associated factor 2 (TRAF2) | 0.817 | 0.773 | 0.713 | 0.465 |
| NM_004001 | Fc fragment of IgG, low affinity IIb, receptor for (CD32) (FCGR2B) | 0.581 | 0.569 | 0.467 | — - |
| NM_001100 | Actin, alpha 1, skeletal muscle (ACTA1) | 0.368 | 0.266 | 0.139 | — - |
| NM_001615 | Actin, gamma 2, smooth muscle, enteric (ACTG2) | 0.419 | 0.229 | 0.191 | - — |
| NM_005718 | Actin related protein 2/3 complex, subunit 4 (20 ku) (ARPC4) | 0.461 | 0.362 | 0.370 | 0.611 |
| NM_020040 | Tubulin, beta polypeptide 4, member Q (TUBB4Q) | 0.462 | 0.345 | 0.335 | 0.573 |
| NM_006009 | Tubulin, alpha 3 (TUBA3) | 1.062 | 0.710 | 0.484 | 0.885 |
| NM_001665 | ras homolog gene family, member G (rho G) (ARHG) | 0.590 | 0.445 | 0.524 | 0.590 |
| AB055890 | c-lbc mRNA for guanine nucleotide exchange factor Lbc | 1.065 | 0.315 | 0.701 | 0.433 |
| NM_006400 | Dynactin 2 (p50) (DCTN2) | 0.865 | 0.361 | 0.334 | — - |
| NM_005428 | vav 1 oncogene (VAV1) | 0.510 | 0.421 | 0.387 | — - |
| NM_005993 | Tubulin-specific chaperone d (TBCD) | 0.770 | 0.485 | 0.524 | — - |
| NM_001747 | Capping protein (actin filament), gelsolin-like (CAPG) | 0.512 | 0.213 | 0.191 | 0.323 |
| GeneBank ID | Gene product | Microarray ratio | Northern blot ratio | ||||||
| 2 h | 6 h | 10 h | 24 h | 2 h | 6 h | 10 h | 24 h | ||
| NM_002817 | PSMD13 | 0.445 | 0.122 | 0.333 | 0.402 | 0.07 | 0.71 | 0.25 | 0.33 |
| NM_004517 | ILK | 0.717 | 0.218 | 0.245 | 0.454 | 0.57 | 0.19 | 0.43 | 0.15 |
Helicobacter pylori has found its own way to thrive within host cells, despite the presence of a well-functioning immune system. In the present study, differential expression of macrophage upon stimulation of VacA gives clear evidence that this vacuolating cytotoxin has evolved with various tricks to escape from the innate and adaptive immunity of the host.
Recognition of pathogen-infected host cells by effector T lymphocytes requires intracellular processing of microbial antigens and their presentation on the surface of the antigen-presenting cells (APCs) in association with MHC molecules. To avoid immune recognition, many kinds of microorganisms have evolved mechanisms which interfere with antigen presentation pathways. These mechanisms may be pivotal, especially for those pathogens causing persistent infections. The present differential expressions provide clear evidence that VacA downregulates the expression of MHC class Iand class II molecules in macrophage. Thus, specific T lymphocytes may not be activated, resulting in the evasion of H pylori from the host’s immune response and colonization of the bacteria in gastric mucosa persistently, remaining “invisible” to both CD8+ and CD4+ T cells. Interference with the class II antigen presentation pathway by Listeria monocytogenes has been shown to result in a reduced proliferation of CD4+ T lymphocytes in response to heterologous antigens[6]. Interactions between MHC class I and human immunodeficiency virus (HIV) resulted in down-regulation of MHC-Isurface expression, contributing to pathogenesis by suppressing the host’s immune response[7]. The role of MHC class I and class II-restricted functions in H pylori infection and immunity upon oral immunization has also been examined in vivo. It was found that experimental challenge with H pylori resulted in significantly greater colonization in MHC class I and class II mutant mice than in wild-type mice[8]. MHC class II-deficient mice were unable to respond to oral antigenic stimulation and remained persistently infected with H pylori[8].
Nitric oxide (NO) is an important mediator of biological processes including inflammatory response. It is synthesized from L-arginine by a family of nitric oxide synthases (NOS), in which, endothelial NOS (eNOS) is a constitutively expressed isoform present in vascular endothelial cells, cardiac myocytes, and blood platelets[9]. eNOS interacting protein (NOSIP) is a 34 ku protein specifically binds to the carboxylterminal region of the human eNOS oxygenase domain, overexpression of which in eNOS-expressing cells has been demonstrated to inhibit NO sythase activity[10]. Thus, down-regulation of this protein upon stimulation of VacA may relieve such inhibition, leading to more NO production. The original concept that the small quantities of NO generated in a pulsatile fashion by constitutive eNOS mainly fulfil regulatory functions required for normal homeostatic function of the vasculature, while the high amount of NO produced by inducible NOS (iNOS) exerts antimicrobial and cytotoxic effects in the immune system, has recently been modified[11]. The expression of eNOS is not restricted to endothelial cells, as it has been found to be present in monocytes/macrophages and in B and T lymphocytes[12]. eNOS can also assume typical immunological functions previously assigned to iNOS, such as the induction of apoptotic cell death and the control of viruses[11]. NO endows macrophages with cytostatic or cytotoxic activity against microbes and tumor cells[13]. Nevertheless, more and more evidences have demonstrated a role for NO in the induction of immunosuppression by inhibiting T-cell proliferation during G1/S transition[14-16]. In mouse models of T-cell-mediated autoimmunity, such as myelin antigen-induced EAE, the disease was exacerbated by genetic deletion of iNOS, indicating that NO suppressed T-cell-mediated immunity in vivo[17]. In addition, NO induces apoptosis, which, the ubiquitin/proteasome and NF-κB pathway have been determined to be involved in[18].
In eukaryotic cells, degradation of many proteins involves their initial modification by conjugation of ubiquitin (Ub). Ubiquinated proteins are rapidly degraded by the 26S proteasome[19]. NO can inhibit the activities of the 20S and 26S proteasomes, providing a likely mechanism for the accumulation of NO-induced pro-apoptotic proteins p53 and Bax, the substrates of the ubiquitin/proteasome system[18]. Apart from the indirect effect via NO functioning, the mRNA expression of proteasomes shows a significant downregulation as the result of being directly stimulated by VacA. Ub/proteasome pathway can catalyze the proteolytic processing of inactive 105 ku NF-κB precursor into 50 ku subunit. p50 is then maintained in the cytosol conjugated with the p65 subunit in an inactive complex bound to IκB. In addition, Ub/proteasome is involved in proteolytic digestion of IκB, which is required for NF-κB activation[19]. Therefore, decreased proteasome activity should inhibit the proteolysis of NF-κB precursor and inhibit IκB degradation, thus blocking NF-κB activation. Another involved protein, integrin-linked kinase (ILK), which has been determined to upregulate NF-κB activity[20], also shows decreased expression in this study. It might be the result of VacA induction, and moreover, ILK mRNA expression was found to be downregulated by NO[21], corresponding to the above speculation that more NO may be produced due to downregulation of NOSIP. NF-κB is known to be important to cell survival. Fibroblasts and macrophages from Rel A (p65 subunit of NF-κB) (-/-) mice were sensitive to TNF-α-induced cell death, and reintroduction of Rel A enhanced cell survival[22]. Activation of NF-κB is required for inflammatory cytokine release by macrophages during infection[23]. Consequently, inhibition of NF-κB activation may be responsible for decreased cytokine release from macrophages and the resulting immunosuppression. Additionally, because ILK is an apoptosis suppressor[24], decreased production of this protein in macrophage may accelerate apoptosis of the cell that plays important roles in innate host defense and antigen presentation, leading to the evasion from host immunity against H pylori.
The capability of degrading proteins by the proteasome accounts for another important function to generate peptides presented on MHC-class I molecules to circulating lymphocytes. The presentation of these peptides enables the immune system to screen for and destroy cells expressing unusual polypeptides[19]. Selective preteasome inhibitors were determined to prevent MHC-class I presentation of the antigenic peptide[25]. Moreover, LMP2 and LMP7, two subunits of the proteasome, were found to be encoded in the major histocompatibility complex (MHC)[26-28]. The experiment using specific antibodies against LMP2 and LMP7 showed that they were co-expressed with MHC-class I molecules[29]. The levels of MHC-class I expression were shown to coincide perfectly with the LMP levels in different tissues, corresponding to the result in the present study, which shows simultaneous downregulation of MHC-class I and LMP7.
MSP, also known as HGF-like protein, is a serum protein belonging to the plasminogen-related growth factor family. It was originally discovered by Leonard and Skeel as a serum protein that stimulates shape change, movement, chemotaxis and phagocytosis of mouse peritoneal resident macrophages[30,31]. The other important effect of MSP on macrophages was to inhibit endotoxin- or cytokine-stimulated NO production[32]. Thereby, the decreased expression of MSP may contribute to deactivating macrophage and producing more NO, which, as described above, functions as an immunosuppressor.
The early growth response 1 (EGR1), a zinc-finger transcription factor that was shown to be significantly upregulated by 2 h postinfection in the present study, has been determined to induce downregulation of copper-zinc superoxide dismutase and manganese superoxide dismutase and stimulate the generation of reactive oxygen species (ROS) via the NADH/NADPH-oxidase system[33]. In addition to NO, ROS produced by NADPH oxidase also have the ability to inhibit the proliferation of lymphocytes by a mechanism that suppressor macrophages impair the proliferative response of T lymphocytes to antigens or mitogens[11]. Otsuji et al.[34] demonstrated that the oxidative stress from tumor-derived macrophages mediated the decrease of CD3ζ chain within T cells, which suppressed the antigen-specific T-cell responses. Pre-treatment of CTL or NK cells with nontoxic concentrations of H2O2 severely reduced their cytotoxic activity, leading to the speculation that macrophage-derived reactive oxygen metabolites contribute directly to alterations in signal transducing molecules of T cells and NK cells and to the mechanism of immunosuppression[35]. Furthermore, the defective expression of CD3ζ on lymphocytes has been related to some kind of carcinoma, including gastric adenocarcinoma[36,37].
From Table 1, we may find up-regulation of MRC OX-2 (the antigen identified by monoclonal antibody, MOX2), a broadly expressed membrane glycoprotein, which has been shown to be important for regulation of the macrophage lineage. In the OX-2-deficient spleens, the number of macrophages was nearly twice the number of those in normal spleens[38], implying a role of OX-2 in suppressing the activation of macrophage. The immunosuppression effects of OX-2 could be further determined by many other studies. For example, several studies reported that increased expression of OX-2 in mice receiving renal allografts was associated with immunosuppression leading to increased graft survival, along with the polarization of cytokine production to type 2 cytokines in lymphocytes harvested from the transplanted animals[39-41]. Furthermore, infusion of a mAb to OX-2 blocks both the increased graft survival and the altered cytokine production[42]. All these data make clear that upregulation of OX-2 does favor to the immune evasion of H pylori.
Table 1 also demonstrates significant downregulation of many cytoskeleton-related gene expression. For example, RhoG is a member of the Rho family of GTPases, which signals to actin assembly during phagocytosis[43]. Vav1 serves as a guanine nucleotide exchange factor (GEF) for Rho proteins, and establishes an essential and direct link between receptors with intrinsic or associated tyrosine kinase activity and the mitogenic and cytoskeletal pathways regulated by Rho proteins[44]. Cytoskeleton has been known to be an important structural basis for phagocytosis since cytochalasin B, a toxin that blocks actin polymerization, was shown to inhibit uptake of IgG-coated erythrocytes by mouse macrophages[45]. Phagocytosis is a process by which macrophages and leukocytes could ingest microbial pathogens to accomplish two essential immune functions, i.e., to initiate the microbial death pathway, and to direct antigens to both MHC I and MHC II compartments. That is to say, phagocytosis serves not only as an innate immune effector but as a bridge between the innate and acquired immune responses[1]. Thus, to destruct cytoskeleton may be another trick of VacA to evade host immune response.
In conclusion, we have shown that H pylori VacA induces the alteration of a series of genes related to immune evasion in macrophage, which ultimately establishes a state of host-microbial equilibrium. Some of these genes are for the first time made an association with VacA stimulation. Further investigations of the previously uncharacterized genes should be made to help us see through the underlying mechanisms utilized by H pylori to escape host immunity.
Edited by Kumar M and Xu FM
| 1. | Ibraghimov A, Pappo J. The immune response against Helicobacter pylori--a direct linkage to the development of gastroduodenal disease. Microbes Infect. 2000;2:1073-1077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 2. | Knipp U, Birkholz S, Kaup W, Opferkuch W. Partial characterization of a cell proliferation-inhibiting protein produced by Helicobacter pylori. Infect Immun. 1996;64:3491-3496. [PubMed] |
| 3. | Telford JL, Ghiara P, Dell'Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce MF, Censini S, Covacci A, Xiang Z. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994;179:1653-1658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 445] [Cited by in RCA: 435] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 4. | Molinari M, Salio M, Galli C, Norais N, Rappuoli R, Lanzavecchia A, Montecucco C. Selective inhibition of Ii-dependent antigen presentation by Helicobacter pylori toxin VacA. J Exp Med. 1998;187:135-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 208] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 5. | Yuan JP, Li T, Shi XD, Hu BY, Yang GZ, Tong SQ, Guo XK. Deletion of Helicobacter pylori vacuolating cytotoxin gene by introduction of directed mutagenesis. World J Gastroenterol. 2003;9:2251-2257. [PubMed] |
| 6. | Leyva-Cobian F, Unanue ER. Intracellular interference with antigen presentation. J Immunol. 1988;141:1445-1450. [PubMed] |
| 7. | Kamp W, Breij EC, Nottet HS, Berk MB. Interactions between major histocompatibility complex class II surface expression and HIV: implications for pathogenesis. Eur J Clin Invest. 2001;31:984-991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 8. | Pappo J, Torrey D, Castriotta L, Savinainen A, Kabok Z, Ibraghimov A. Helicobacter pylori infection in immunized mice lacking major histocompatibility complex class I and class II functions. Infect Immun. 1999;67:337-341. [PubMed] |
| 9. | Michel T, Feron O. Nitric oxide synthases: which, where, how, and why. J Clin Invest. 1997;100:2146-2152. [PubMed] |
| 10. | Dedio J, König P, Wohlfart P, Schroeder C, Kummer W, Müller-Esterl W. NOSIP, a novel modulator of endothelial nitric oxide synthase activity. FASEB J. 2001;15:79-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 138] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 11. | Bogdan C, Röllinghoff M, Diefenbach A. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr Opin Immunol. 2000;12:64-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 675] [Cited by in RCA: 639] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 12. | Reiling N, Kröncke R, Ulmer AJ, Gerdes J, Flad HD, Hauschildt S. Nitric oxide synthase: expression of the endothelial, Ca2+/calmodulin-dependent isoform in human B and T lymphocytes. Eur J Immunol. 1996;26:511-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3127] [Cited by in RCA: 3071] [Article Influence: 109.7] [Reference Citation Analysis (0)] |
| 14. | Albina JE, Abate JA, Henry WL. Nitric oxide production is required for murine resident peritoneal macrophages to suppress mitogen-stimulated T cell proliferation. Role of IFN-gamma in the induction of the nitric oxide-synthesizing pathway. J Immunol. 1991;147:144-148. [PubMed] |
| 15. | Eisenstein TK, Huang D, Meissler JJ, al-Ramadi B. Macrophage nitric oxide mediates immunosuppression in infectious inflammation. Immunobiology. 1994;191:493-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Liew FY. Regulation of lymphocyte functions by nitric oxide. Curr Opin Immunol. 1995;7:396-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 134] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | van der Veen RC. Nitric oxide and T helper cell immunity. Int Immunopharmacol. 2001;1:1491-1500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 94] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 18. | Glockzin S, von Knethen A, Scheffner M, Brüne B. Activation of the cell death program by nitric oxide involves inhibition of the proteasome. J Biol Chem. 1999;274:19581-19586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 87] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1836] [Cited by in RCA: 1843] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 20. | Tan C, Mui A, Dedhar S. Integrin-linked kinase regulates inducible nitric oxide synthase and cyclooxygenase-2 expression in an NF-kappa B-dependent manner. J Biol Chem. 2002;277:3109-3116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Beck KF, Walpen S, Eberhardt W, Pfeilschifter J. Downregulation of integrin-linked kinase mRNA expression by nitric oxide in rat glomerular mesangial cells. Life Sci. 2001;69:2945-2955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 22. | Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2509] [Cited by in RCA: 2484] [Article Influence: 85.7] [Reference Citation Analysis (0)] |
| 23. | Baeuerle PA, Henkel T. Function and activation of NF-kappa B in the immune system. Annu Rev Immunol. 1994;12:141-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3598] [Cited by in RCA: 3639] [Article Influence: 117.4] [Reference Citation Analysis (1)] |
| 24. | Attwell S, Roskelley C, Dedhar S. The integrin-linked kinase (ILK) suppresses anoikis. Oncogene. 2000;19:3811-3815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 180] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 25. | Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1889] [Cited by in RCA: 1968] [Article Influence: 63.5] [Reference Citation Analysis (0)] |
| 26. | Brown MG, Driscoll J, Monaco JJ. Structural and serological similarity of MHC-linked LMP and proteasome (multicatalytic proteinase) complexes. Nature. 1991;353:355-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 232] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 27. | Martinez CK, Monaco JJ. Homology of proteasome subunits to a major histocompatibility complex-linked LMP gene. Nature. 1991;353:664-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 200] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 28. | Yang Y, Waters JB, Früh K, Peterson PA. Proteasomes are regulated by interferon gamma: implications for antigen processing. Proc Natl Acad Sci USA. 1992;89:4928-4932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 141] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 29. | Früh K, Yang Y, Arnold D, Chambers J, Wu L, Waters JB, Spies T, Peterson PA. Alternative exon usage and processing of the major histocompatibility complex-encoded proteasome subunits. J Biol Chem. 1992;267:22131-22140. [PubMed] |
| 30. | Leonard EJ, Skeel A. A serum protein that stimulates macrophage movement, chemotaxis and spreading. Exp Cell Res. 1976;102:434-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 72] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 31. | Leonard EJ, Skeel AH. Isolation of macrophage stimulating protein (MSP) from human serum. Exp Cell Res. 1978;114:117-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 32. | Wang MH, Cox GW, Yoshimura T, Sheffler LA, Skeel A, Leonard EJ. Macrophage-stimulating protein inhibits induction of nitric oxide production by endotoxin- or cytokine-stimulated mouse macrophages. J Biol Chem. 1994;269:14027-14031. [PubMed] |
| 33. | Bek MJ, Reinhardt HC, Fischer KG, Hirsch JR, Hupfer C, Dayal E, Pavenstadt H. Up-regulation of early growth response gene-1 via the CXCR3 receptor induces reactive oxygen species and in-hibits Na /K -ATPase activity in an immortalized human proxi-mal tubule cell line. J Immunol. 2003;170:931-940. [RCA] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 34. | Otsuji M, Kimura Y, Aoe T, Okamoto Y, Saito T. Oxidative stress by tumor-derived macrophages suppresses the expression of CD3 zeta chain of T-cell receptor complex and antigen-specific T-cell responses. Proc Natl Acad Sci USA. 1996;93:13119-13124. [RCA] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 246] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 35. | Kono K, Salazar-Onfray F, Petersson M, Hansson J, Masucci G, Wasserman K, Nakazawa T, Anderson P, Kiessling R. Hydrogen peroxide secreted by tumor-derived macrophages down-modulates signal-transducing zeta molecules and inhibits tumor-specific T cell-and natural killer cell-mediated cytotoxicity. Eur J Immunol. 1996;26:1308-1313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 245] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 36. | Kono K, Ichihara F, Iizuka H, Sekikawa T, Matsumoto Y. Expression of signal transducing T-cell receptor zeta molecules after adoptive immunotherapy in patients with gastric and colon cancer. Int J Cancer. 1998;78:301-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 37. | Takahashi A, Kono K, Amemiya H, Iizuka H, Fujii H, Matsumoto Y. Elevated caspase-3 activity in peripheral blood T cells coexists with increased degree of T-cell apoptosis and down-regulation of TCR zeta molecules in patients with gastric cancer. Clin Cancer Res. 2001;7:74-80. [PubMed] |
| 38. | Hoek RM, Ruuls SR, Murphy CA, Wright GJ, Goddard R, Zurawski SM, Blom B, Homola ME, Streit WJ, Brown MH. Down-regulation of the macrophage lineage through interaction with OX2 (CD200). Science. 2000;290:1768-1771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 749] [Cited by in RCA: 776] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 39. | Gorczynski L, Chen Z, Hu J, Kai Y, Lei J, Ramakrishna V, Gorczynski RM. Evidence that an OX-2-positive cell can inhibit the stimulation of type 1 cytokine production by bone marrow-derived B7-1 (and B7-2)-positive dendritic cells. J Immunol. 1999;162:774-781. [PubMed] |
| 40. | Gorczynski RM, Yu K, Clark D. Receptor engagement on cells expressing a ligand for the tolerance-inducing molecule OX2 in-duces an immunoregulatory population that inhibits alloreactivity in vitro and in vivo. J Immunol. 2000;165:4854-4860. [RCA] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 59] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 41. | Gorczynski RM, Chen Z, Hu J, Kai Y, Lei J. Evidence of a role for CD200 in regulation of immune rejection of leukaemic tumour cells in C57BL/6 mice. Clin Exp Immunol. 2001;126:220-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 42. | Gorczynski RM, Cattral MS, Chen Z, Hu J, Lei J, Min WP, Yu G, Ni J. An immunoadhesin incorporating the molecule OX-2 is a potent immunosuppressant that prolongs allo- and xenograft survival. J Immunol. 1999;163:1654-1660. [PubMed] |
| 43. | Vigorito E, Billadeu DD, Savoy D, McAdam S, Doody G, Fort P, Turner M. RhoG regulates gene expression and the actin cytoskeleton in lymphocytes. Oncogene. 2003;22:330-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 44. | Bustelo XR. Regulatory and signaling properties of the Vav family. Mol Cell Biol. 2000;20:1461-1477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 421] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 45. | Kaplan G. Differences in the mode of phagocytosis with Fc and C3 receptors in macrophages. Scand J Immunol. 1977;6:797-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 156] [Article Influence: 3.3] [Reference Citation Analysis (0)] |