Published online May 15, 2004. doi: 10.3748/wjg.v10.i10.1508
Revised: May 12, 2003
Accepted: May 19, 2003
Published online: May 15, 2004
AIM: To investigate the relationship between gallbladder stone disease (GSD) and single nucleotide polymorphisms of cholesterol 7α-hydroxylase (CYP7A) gene promoter, apolipoprotein (APO) B gene exon 26, APOE gene exon 4 or microsatellite polymorphism of low density lipoprotein receptor (LDLR) gene exon 18.
METHODS: Genotypes of CYP7A, APOB, APOE and LDLR genes were determined in 105 patients with GSD diagnosed by B-mode ultrasonography and 274 control subjects. Serum lipids were analyzed with HITACHI 7060 automaic biochemical analyzer.
RESULTS: Body mass index (BMI) was significantly higher in patients with GSD (24.47 ± 3.09) than in controls (23.50 ± 2.16). Plasma total cholesterol was lower in patients with GSD (4.66 ± 0.92 mmol/L) than in controls (4.91 ± 0.96 mmol/L), P < 0.01 after adjusted for age, sex and BMI. The significantly higher frequency of A allele of CYP7A gene polymorphism and X+ allele of APOB gene polymorphism was seen in GSD patients. Percentages of A allele in patients and controls were 62.86% and 54.38% (P < 0.05) and those of X+ allele 8.57% and 4.01% (P < 0.01). Subjects with A allele had significantly lower plasma total cholesterol and LDL cholesterol than subjects with CC homozygote. In a multiple variable logistic regression model, the BMI (OR = 1.13, 95%CI: 1.05-1.22), A allele (OR = 1.48, 95%CI: 1.05-2.09) and X+ allele (OR = 2.28, 95%CI: 1.14-4.59) were positively associated with GSD (P < 0.05). Plasma total cholesterol (OR = 0.69, 95%CI: 0.64-0.74) was negatively related to GSD (P < 0.05).
CONCLUSION: With an association analysis, it was determined that A allele of CYP7A gene and X+ allele of APOB gene might be considered as risk genes for GSD. These alleles are related with differences of serum lipids among subjects. Multiple-variable logistic regression model analysis showed that besides BMI, GSD was affected by polygenetic factors. But the mechanism for these two alleles responsible for GSD requires further investigations.
- Citation: Jiang ZY, Han TQ, Suo GJ, Feng DX, Chen S, Cai XX, Jiang ZH, Shang J, Zhang Y, Jiang Y, Zhang SD. Polymorphisms at cholesterol 7α-hydroxylase, apolipoproteins B and E and low density lipoprotein receptor genes in patients with gallbladder stone disease. World J Gastroenterol 2004; 10(10): 1508-1512
- URL: https://www.wjgnet.com/1007-9327/full/v10/i10/1508.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i10.1508
Gallbladder stone disease (GSD) is prevalent in China with a gradually increasing incidence. Studies on the pathogenesis of GSD have demonstrated that supersaturation of biliary cholesterol caused by excessive biliary cholesterol and/or decreased bile acid is the requisite for the formation of gallstones[1]. Both the cholesterol secreted into bile and the bile acids converted from cholesterol in the liver are involved in regulating cholesterol homeostasis. It was shown in studies[2-4] that genetic variations might affect gallstone formation. In 1995, Khajuana et al[4] reported a murine lithogenic gene, Lith 1, as a possible regulator of hepatic cholesterol synthesis. The primary Lith phenotype was considered to induce secondary events characterized by multiple enzyme alterations which increase available cholesterol and supply the sterol to hepatocytes for hypersecretion into bile[5]. Additional quantitative traits linkage analysis[6] maps other Lith genes on murine chromosomes 6, 7, 8, 10, 19 and X to confirm the polygenic mode of inheritance. A few studies[7-10] have focused on the relationship between human GSD and certain genetic factors contributing to cholesterol metabolism.
Cholesterol 7α-hydroxylase (CYP7A, EC 1.14.13.17), a cytochrome P-450 enzyme, is the rate limiting enzyme of hepatic bile acid synthesis, with its activity regulated by bile acids, cholesterol and hormones[11]. Although the amino acid sequence of CYP7A between species is highly homologous (80%-90% sequence identity), species respond differently to diet cholesterol[12]. As compared with control subjects, the activity of CYP7A varied in patients with gallstones[13-15], and diminished or elevated patterns were observed. The heterogeneity of activities of CYP7A in patients with GSD may be related to CYP7A polymorphisms. A linkage of A-204C single nucleotide polymorphism of the CYP7A gene promoter with plasma low density lipoprotein (LDL) cholesterol was found in recent studies of nuclear families[16] and within the general population[17]. However, the polymorphism for patients with GSD has never been studied. Apolipoprotein (APO) E[18] is an extremely efficient ligand for the LDLR and is the determinant for receptor-mediated catabolism of all APOE containing lipoproteins. The polymorphism of APOE is controlled by three alleles in exon 4, namely ε2, ε3 and ε4. The physiological importance of APOE exhibits in disorders of lipoprotein metabolism such as atherosclerosis[19] as well as in Alzheimer’s disease which is not obviously related to lipoprotein metabolism[20]. The role of APOE has also been examined in relation to GSD[7,9]. The APOE4 allele is associated with high cholesterol content in gallstones[7,9], faster crystallization[7] and frequent stone recurrence after lithotripsy[21]. The LDLR on the surface of hepatocytes plays an important role in cholesterol homeostasis in humans[22]. The receptor can recognize APOB or APOE containing lipoproteins such as LDL and high-density lipoprotein (HDL) with different affinities mediating the absorption of plasma lipoproteins. There are many polymorphic sites on this gene and some of those are related to plasma cholesterol metabolism[23].
In the present study, we analyzed the polymorphism of A-204C of CYP7A gene promoter, APOE exon 4 and microsatellite polymorphism of LDLR gene exon 18. Their relationships with asymptomatic GSD on the Chinese Han population were examined. The association of Xba I polymorphism on APOB gene exon 26 with GSD, shown in our previous study[10], was also evaluated using multiple regression analysis.
A total of 379 subjects were recruited for this study from February to May 1998. Patients in this study consisted of 78 males and 27 females with stones and/or cholesterol crystals in their gallbladder. None of the patients had previous onset of cholecystitis defined as colic, fever with chills or jaundice. Two hundred and seventy-four healthy subjects (184 males and 90 females) with normal liver, kidney and endocrine function were included as controls. The mean ages of patients and controls were 47.53 and 47.94 years, respectively. After a 12-h fast, the participants received B-mode ultrasonography with Aloka 500/SSD equipped with a transducer of 3.5 mHz. A total of 10 mL venous blood was extracted and half of it was immediately mixed with ACD anti-coagulants containing citric acid, sodium citric acid and glucose for DNA extraction. The other half of this sample was prepared for biochemical analysis. Body mass index (BMI) was calculated by weight/height2 (kg/m2). All subjects gave informed consent to participate in this study which was approved by the Ethical Committee of Ruijin Hospital, Shanghai Second Medical University.
Plasma total cholesterol, triglyceride, HDL cholesterol, LDL cholesterol, APO AI and APO B were assayed by commercially available kits (Boehringer Mannheim GmBH, Mannheim, Germany) on an automatic analyzer (HITACHI 7060, Hitachi Koki Co. Ltd., Hitachinaka City, Japan).
Genomic DNA was extracted from leukocytes using a method provided by GIBCO-BRL DNA extraction kit (Cat: #28350-015, GIBCO-BRL, Gaithersburg, MD, USA). The fragments containing target polymorphic sequences of CYP7A, APOB, APOE and LDLR genes were amplified using polymerase chain reaction (PCR) on PTC-200 Peltier Thermal Cycler (MJ Research Inc, MA, USA). Primers and the conditions of PCR are listed in Table 1. For the single nucleotide polymorphisms of CYP7A, APOB and APOE, the products of PCR were each digested by restriction enzymes (New England Biolabs Inc., Beverly, MA, USA). The enzymes are indicated in Table 1. The digested PCR products were electrophoresed in agarose gel; then stained with ethidium bromide and visualized under ultraviolet light. A 968-bp fragment containing A-204C polymorphism of CYP7A gene was digested with restrictive enzyme Bsa I (New England Biolabs Inc., Beverly, MA, USA) and electrophoresed on 10 g/L agarose gel and stained with ethidium bromide. The band with a cutting site was designated as C allele and that without as A allele. A 244-bp fragment containing APOE exon 4 gene was digested with Hha I followed by electrophoresis on 100 g/L polyacrylamide gel and then stained with silver. The genotypes were determined from the pattern of restrictive fragments on the gel as described in detail by Hixson and Vernier[24]. The bands representing the genotype of microsatellite polymorphism of LDLR gene exon 18 were obtained from direct electrophoresis on 100 g/L PAGE (100 v, 2.5 h) and stained with silver. The genotype of LDLR gene was determined for the bands with 106-bp as A allele (7 repeats of TA), 108-bp as B allele (8 repeats of TA) or 112-bp as C allele (10 repeats of TA)[25]. Genotyping of APOB gene was performed as previously described by Han et al[10].
| Genes | Primers | Annealing temp | RE |
| CYP7A[16,17] | 5’TGGTAGGTAAATTATTAATAGATGT3’ | ||
| 5’AAATTAAATGGATGAATCAAAGAGC3’ | 61°C | Bsa I | |
| apo B[10] | 5’GGAGACTATTCAGAAGCTAA3’ | ||
| 5’GAAGAGCCTGAAGACTGACT3’ | 60°C | Xba I | |
| apo E[24] | 5’ACAGAATTCGCCCCGGCCTGGTACAC3’ | ||
| 5’TAAGCTTGGCACGGCTGTCCAAGGA3’ | 60°C | Hha I | |
| LDL receptor[25] | 5’CACTTTGTATATTGGTTGAAACTGT3’ | ||
| 5’CACTGAACAAATACAGCAACCAGGG3’ | 62°C |
The results were expressed as means ± SD. The differences in concentrations of lipids between patients and controls and those among genotypes were calculated using Student’s t test. Statistical analysis was performed using the statistical software package SAS 6.12 for Windows (SAS Institute Inc., Cary, NC, USA). SAS GLM procedure was used to compare the concentrations of lipids between patients and controls after adjustment for sex, age and BMI. Frequencies of alleles between patients and controls were evaluated for statistical significance using Chi-square test. A multivariate model was used to predict the relative odds of GSD with all the variables by multiple logistic regressions. Logistic regression coefficients and standard errors were calculated to determine the estimates of odds ratio (OR) and 95% confidence intervals (CI) for significant factors.
Gallbladder stones and/or cholesterol crystals were detected in 105 cases by B-mode ultrasonography. The demographic characteristics and biochemistry are shown in Table 2. BMI was significantly higher in the patients than in the controls (24.47 ± 3.09 vs 23.50 ± 2.16, P < 0.01). Concentrations of plasma total cholesterol and APO AI were significantly lower in patients than in controls. Although the concentrations of HDL cholesterol and LDL cholesterol were slightly lower in patients than in controls, the differences were not significant. Plasma lipid levels varied with sex, age and BMI. We compared the plasma lipids between patients and controls using the analysis of covariance. After adjustment for sex, age and BMI, the plasma total cholesterol and LDL cholesterol as well as APO B were found significantly lower in patients than in controls as shown in Table 2.
| Patients | Controls | P value | P value1 | |
| No. (male/female) | 105 (78/27) | 274 (184/90) | ||
| Age (yr) | 47.53 ± 10.98 | 47.94 ± 12.21 | ||
| BMI (kg/m2) | 24.47 ± 3.09 | 23.50 ± 2.16 | < 0.01 | |
| Triglycerine (mmol/L) | 1.32 ± 1.15 | 1.19 ± 0.82 | ||
| Cholesterol (mmol/L) | 4.66 ± 0.92 | 4.91 ± 0.96 | < 0.05 | < 0.01 |
| HDL (mmol/L) | 1.33 ± 0.36 | 1.40 ± 0.34 | ||
| LDL (mmol/L) | 2.56 ± 0.67 | 2.69 ± 0.73 | < 0.05 | |
| apo A I (g/L) | 1.34 ± 0.21 | 1.39 ± 0.18 | < 0.05 | |
| apo B (g/L) | 1.00 ± 0.22 | 1.03 ± 0.24 | < 0.05 |
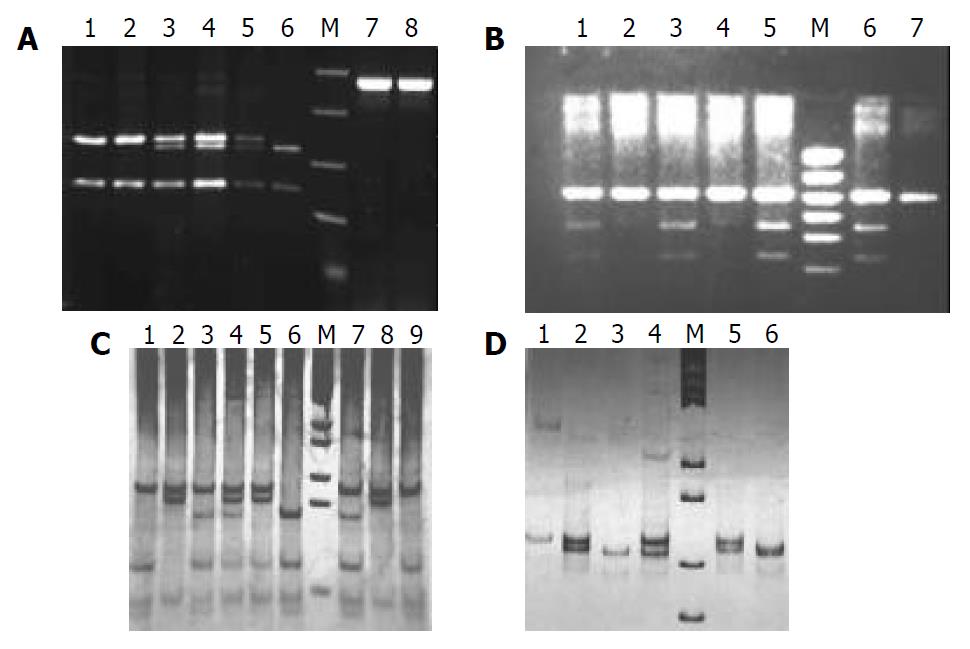
Figure 1 indicates the genotypes of CYP7A, APOB, APOE and LDLR gene polymorphisms. The distributions of genotypes for patients and controls are listed in Table 3. Using Chi-square test, there was a significantly higher frequency of A allele of CYP7A gene and X+ allele of APOB gene observed in patients compared with controls (A allele: 62.86% vs 54.38%, P < 0.05; X+ allele: 8.57% vs 4.01%, P < 0.01, Table 4). There were no significant differences between patients and controls in the polymorphisms of APOE gene and LDLR gene.
| Genes | Genotypes | Patients (%) | Controls (%) |
| CYP7A | AA | 44 (41.90) | 79 (28.83) |
| AC | 44 (41.90) | 140 (51.10) | |
| CC | 17 (16.20) | 55 (20.07) | |
| Apo B | X+/+ | 1 (0.95) | 0 (0) |
| X+/- | 16 (15.24) | 22 (8.03) | |
| X-/- | 88 (83.81) | 252 (91.97) | |
| Apo E | ε2/2 | 0 (0) | 1 (0.36) |
| ε2/3 | 15 (14.29) | 45 (16.42) | |
| ε2/4 | 5 (4.76) | 6 (2.19) | |
| ε3/3 | 73 (69.52) | 183 (66.80) | |
| ε3/4 | 11 (10.48) | 37 (13.50) | |
| ε4/4 | 1 (0.95) | 2 (0.73) | |
| LDL receptor | AA | 41 (39.05) | 90 (32.85) |
| AB | 2 (1.90) | 13 (4.74) | |
| AC | 32 (30.48) | 104 (37.96) | |
| BB | 3 (2.85) | 4 (1.46) | |
| BC | 8 (7.62) | 22 (8.03) | |
| BC | 19 (18.60) | 41 (14.96) |
Table 5 indicates that plasma total cholesterol, LDL cholesterol and APOB were lower in subjects with A allele (AA homozygote or AC heterozygote) than in those without A allele (CC homozygote). The difference was significant only within the control group or within the group combining patients and controls, but not in the patients only group. Within each genotype, the difference of lipid concentration was incongruent between patients and controls. In subjects with A allele, the LDL cholesterol was significantly higher in patients than in controls, while in subjects without A allele, patients had significantly lower concentrations of HDL cholesterol and APOAI.
| Patients | Controls | All | ||||
| AA/AC | CC | AA/AC | CC | AA/AC | CC | |
| No. | 88 | 17 | 219 | 55 | 307 | 72 |
| Triglyceride (mmol/L) | 1.25 ± 0.99 | 1.70 ± 7.77 | 1.15 ± 0.82 | 1.36 ± 0.81 | 1.18 ± 0.87 | 1.44 ± 1.11 |
| Cholesterol (mmol/L) | 4.64 ± 0.95 | 4.78 ± 0.79 | 4.84 ± 0.94c | 5.16 ± 0.99c | 4.78 ± 0.95g | 5.07 ± 0.95g |
| HDL (mmol/L) | 1.32 ± 0.37a | 1.35 ± 0.32 | 1.42 ± 0.34a | 1.33 ± 0.72 | 1.39 ± 0.35 | 1.33 ± 0.32 |
| LDL (mmol/L) | 2.56 ± 0.70 | 2.59 ± 0.45e | 2.63 ± 0.72c | 2.91 ± 0.72e,c | 2.61 ± 0.71g | 2.83 ± 0.67g |
| Apo AI (g/L) | 1.34 ± 0.22a | 1.36 ± 0.15 | 1.39 ± 0.18a | 1.36 ± 0.20 | 1.38 ± 0.19 | 1.36 ± 0.18 |
| Apo B (g/L) | 1.00 ± 0.23 | 1.02 ± 0.15 | 1.01 ± 0.24c | 1.10 ± 0.23c | 1.01 ± 0.23g | 1.08 ± 0.22g |
A multiple variable logistic regression analysis was performed to compare the effects of both genetic factors and other quantitative variables. In Table 6, only CYP7A (OR = 1.48, P < 0.05) and APOB (OR = 2.28, P < 0.05) gene polymorphism, plasma cholesterol (OR = 0.69, P < 0.01) and BMI (OR = 1.13, P < 0.05) were correlated with GSD (Table 6).
Epidemiological studies have stressed the relationship between plasma lipid concentration and GSD. The present study indicated that patients with GSD had lower plasma cholesterol, even after adjustment for sex, age and BMI. Our results were consistent with those of Juvonen[7], Scragg[26] and Attili[27], although there were unchanged results[28] or only lowered plasma cholesterol levels in women[29] reported by other authors. The plasma lipid concentrations varied in populations due to diet habits, genetic factors, ethnicity, etc. This explains why previous studies remain controversial. Whether changes in plasma lipid concentration are major factors inducing GSD or represent the end stage of gallstone formation is still questionable. The mechanism for the change in plasma lipid concentrations that increases the risk for GSD is also unclear. However, epidemiological studies indicate that genetic predisposition has been confirmed to have a close relation with GSD. Our present study on the Chinese Han population verified that GSD was genetically controlled by polygenetic factors. A-204C polymorphism at CYP7A gene promoter and Xba I polymorphism at APOB gene may be susceptible genes linked to GSD.
Our previous study on the APOB gene Xba I polymorphism revealed that X+ allele was associated with a higher incidence of GSD[10]. The relationship between GSD and the other three polymorphic sites, A-204C polymorphism of CYP7A, Hha I polymorphism of APOE and a dinucleotide repeat microsatellite polymorphism of LDLR, was also studied by our group. Only did CYP7A gene polymorphism seem to be related to GSD. We found a significantly higher frequency of A allele in patients than in controls (0.62 vs 0.54, P < 0.05). The A-204C polymorphism located 204 bp upstream of the transcription start site of CYP7A[17]. This single nucleotide polymorphism[16], at which site C replaced A, created a cutting site for the Bsa I restrictive enzyme. The frequency of A allele of CYP7A gene was 0.58[16] in the Caucasian population and 0.60 in Framingham families[17] which was slightly higher than that in the Chinese Han population. Analysis of plasma lipid levels revealed that plasma LDL cholesterol concentrations were associated with A-204C polymorphism, similar to the results of Wang et al[16,17] and Couture. Individuals with A allele tended to have lower LDL cholesterol concentrations. This difference was significant in controls, but not in patients in this study. No studies have assayed the activity of hepatic CYP7A among different genotypes up to this time. The mechanism and rationale for A allele to induce low LDL cholesterol are still unknown.
Contrary to our expectation, there was no association between APOE gene and GSD or between LDLR gene and GSD. Since APOE isoforms had different affinities to receptors[22] affecting lipid metabolism, there was a demonstrated relationship between APOE gene polymorphism and various plasma lipids. Its polymorphism was well documented in atherosclerosis[19] and Alzheimer’s disease[20] as well as cholesterol gallstone disease[7,9]. Juvonen [7] studied for the first time the relationship between APOE polymorphism and gallstones. Patients with ε4 allele had higher cholesterol content in stones, rapid cholesterol crystallization and shorter median nucleation time in the Finnish population[7] while Bertomeu[9] found significantly higher ε4 allele frequency in Spanish patients with gallstones. But other studies[9,30,31] had contradictory results. There were reports that median nucleation time[9] was similar in patients between the genotypes and that the cholesterol saturation index[30] was lower in patients with ε4 allele. Our study concluded that there was no significant difference in the frequency of ε4 allele between patients and controls. A similar result on Chinese patients with GSD was reported previously in a study from Sichuan, China[32].
LDLR was important for the absorption of APOB and APOE containing lipoproteins[22]. Herein, we studied a dinucleotide repeat polymorphism. The frequency of these alleles did not differ between the patient and control groups. But we could not exclude the possibility of existence of other polymorphic sites to account for the differences in hepatic LDLR activities that, in turn, would lead to high absorption of plasma cholesterol for biliary secretion.
We analyzed all our variables using a logistic regression model to evaluate the role of genetic factors and quantitative variables. Higher cholesterol concentration in serum may provide a protective factor for GSD since it is negatively related with GSD (OR < 1). The cause of gallstone formation might be due to hepatic overabsorption of cholesterol via receptors such as SRB1[33] or LDLR[22] leading to hypersaturation of biliary cholesterol. Besides BMI and cholesterol, GSD may be controlled by multiple genetic factors such as the APOB gene or CYP7A gene. The OR of the APOB gene polymorphism was 2.28, and CYP7A was 1.48 with P < 0.05. This implies that subjects with A allele of CYP7A gene or X+ allele of APOB gene can easily form gallstones.
The present study determined the relationship between CYP7A or APOB gene polymorphism and GSD. However, the relationship should be investigated further in family pedigrees with GSD. Since gene polymorphisms are heterogeneous among ethnic groups, GSD may be caused by different risk genes among different population. The main cause for GSD is hypersaturation of biliary cholesterol. So all genes (known or yet to be determined) involved in hepatic cholesterol metabolism remain the focus for future studies to discover the primary risk genes for GSD. Once a human genomic map for GSD, similar to murine[34], is completed, it can be useful for early predictive and preventive measures for subjects susceptible to GSD. The genomic map will provide the necessary link in the discovery of effective pharmaceutical agents for treatment.
Edited by Ma JY and Wang XL Proofread by Xu FM
| 1. | Apstein MD, Carey MC. Pathogenesis of cholesterol gallstones: a parsimonious hypothesis. Eur J Clin Invest. 1996;26:343-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 87] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 2. | Miquel JF, Covarrubias C, Villaroel L, Mingrone G, Greco AV, Puglielli L, Carvallo P, Marshall G, Del Pino G, Nervi F. Genetic epidemiology of cholesterol cholelithiasis among Chilean Hispanics, Amerindians, and Maoris. Gastroenterology. 1998;115:937-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 143] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 3. | Duggirala R, Mitchell BD, Blangero J, Stern MP. Genetic determinants of variation in gallbladder disease in the Mexican-American population. Genet Epidemiol. 1999;16:191-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 4. | Khanuja B, Cheah YC, Hunt M, Nishina PM, Wang DQ, Chen HW, Billheimer JT, Carey MC, Paigen B. Lith1, a major gene affecting cholesterol gallstone formation among inbred strains of mice. Proc Natl Acad Sci USA. 1995;92:7729-7733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 158] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 5. | Lammert F, Wang DQ, Paigen B, Carey MC. Phenotypic characterization of Lith genes that determine susceptibility to cholesterol cholelithiasis in inbred mice: integrated activities of hepatic lipid regulatory enzymes. J Lipid Res. 1999;40:2080-2090. [PubMed] |
| 6. | Paigen B, Schork NJ, Svenson KL, Cheah YC, Mu JL, Lammert F, Wang DQ, Bouchard G, Carey MC. Quantitative trait loci mapping for cholesterol gallstones in AKR/J and C57L/J strains of mice. Physiol Genomics. 2000;4:59-65. [PubMed] |
| 7. | Juvonen T, Kervinen K, Kairaluoma MI, Lajunen LH, Kesäniemi YA. Gallstone cholesterol content is related to apolipoprotein E polymorphism. Gastroenterology. 1993;104:1806-1813. [PubMed] |
| 8. | Juvonen T, Savolainen MJ, Kairaluoma MI, Lajunen LH, Humphries SE, Kesäniemi YA. Polymorphisms at the apoB, apoA-I, and cholesteryl ester transfer protein gene loci in patients with gallbladder disease. J Lipid Res. 1995;36:804-812. [PubMed] |
| 9. | Bertomeu A, Ros E, Zambón D, Vela M, Pérez-Ayuso RM, Targarona E, Trías M, Sanllehy C, Casals E, Ribó JM. Apolipoprotein E polymorphism and gallstones. Gastroenterology. 1996;111:1603-1610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 67] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 10. | Han T, Jiang Z, Suo G, Zhang S. Apolipoprotein B-100 gene Xba I polymorphism and cholesterol gallstone disease. Clin Genet. 2000;57:304-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 11. | Einarsson C, Ellis E, Abrahamsson A, Ericzon BG, Bjorkhem I, Axelson M. Bile acid formation in primary human hepatocytes. World J Gastroenterol. 2000;6:522-525. [PubMed] |
| 12. | Xu G, Shneider BL, Shefer S, Nguyen LB, Batta AK, Tint GS, Arrese M, Thevananther S, Ma L, Stengelin S. Ileal bile acid transport regulates bile acid pool, synthesis, and plasma cholesterol levels differently in cholesterol-fed rats and rabbits. J Lipid Res. 2000;41:298-304. [PubMed] |
| 13. | Ito T, Kawata S, Imai Y, Kakimoto H, Trzaskos JM, Matsuzawa Y. Hepatic cholesterol metabolism in patients with cholesterol gallstones: enhanced intracellular transport of cholesterol. Gastroenterology. 1996;110:1619-1627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 14. | Reihnér E, Angelin B, Björkhem I, Einarsson K. Hepatic cholesterol metabolism in cholesterol gallstone disease. J Lipid Res. 1991;32:469-475. [PubMed] |
| 15. | Shoda J, He BF, Tanaka N, Matsuzaki Y, Osuga T, Yamamori S, Miyazaki H, Sjövall J. Increase of deoxycholate in supersaturated bile of patients with cholesterol gallstone disease and its correlation with de novo syntheses of cholesterol and bile acids in liver, gallbladder emptying, and small intestinal transit. Hepatology. 1995;21:1291-1302. [PubMed] |
| 16. | Wang J, Freeman DJ, Grundy SM, Levine DM, Guerra R, Cohen JC. Linkage between cholesterol 7alpha-hydroxylase and high plasma low-density lipoprotein cholesterol concentrations. J Clin Invest. 1998;101:1283-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 102] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Couture P, Otvos JD, Cupples LA, Wilson PW, Schaefer EJ, Ordovas JM. Association of the A-204C polymorphism in the cholesterol 7alpha-hydroxylase gene with variations in plasma low density lipoprotein cholesterol levels in the Framingham Offspring Study. J Lipid Res. 1999;40:1883-1889. [PubMed] |
| 18. | Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2791] [Cited by in RCA: 2919] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 19. | Curtiss LK, Boisvert WA. Apolipoprotein E and atherosclerosis. Curr Opin Lipidol. 2000;11:243-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 177] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 20. | Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977-1981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2712] [Cited by in RCA: 2868] [Article Influence: 89.6] [Reference Citation Analysis (0)] |
| 21. | Portincasa P, van Erpecum KJ, van De Meeberg PC, Dallinga-Thie GM, de Bruin TW, van Berge-Henegouwen GP. Apolipoprotein E4 genotype and gallbladder motility influence speed of gallstone clearance and risk of recurrence after extracorporeal shock-wave lithotripsy. Hepatology. 1996;24:580-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 42] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4078] [Cited by in RCA: 3956] [Article Influence: 101.4] [Reference Citation Analysis (0)] |
| 23. | Pedersen JC, Berg K. Normal DNA polymorphism at the low density lipoprotein receptor (LDLR) locus associated with serum cholesterol level. Clin Genet. 1988;34:306-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 24. | Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31:545-548. [PubMed] |
| 25. | Zuliani G, Hobbs HH. Dinucleotide repeat polymorphism at the 3' end of the LDL receptor gene. Nucleic Acids Res. 1990;18:4300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Scragg RK, Calvert GD, Oliver JR. Plasma lipids and insulin in gall stone disease: a case-control study. Br Med J (Clin Res Ed). 1984;289:521-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 94] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Attili AF, Capocaccia R, Carulli N, Festi D, Roda E, Barbara L, Capocaccia L, Menotti A, Okolicsanyi L, Ricci G. Factors associated with gallstone disease in the MICOL experience. Multicenter Italian Study on Epidemiology of Cholelithiasis. Hepatology. 1997;26:809-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 140] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 28. | Barbara L, Sama C, Morselli Labate AM, Taroni F, Rusticali AG, Festi D, Sapio C, Roda E, Banterle C, Puci A. A population study on the prevalence of gallstone disease: the Sirmione Study. Hepatology. 1987;7:913-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 324] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 29. | Barbara L; GREPCO. The epidemiology of gallstone disease in Rome, Italy. Part II. Factors associated with the disease. Hepatology. 1988;8:907-913. [PubMed] |
| 30. | Van Erpecum KJ, Van Berge-henegouwen GP, Eckhardt ER, Portincasa P, Van De Heijning BJ, Dallinga-Thie GM, Groen AK. Cholesterol crystallization in human gallbladder bile: relation to gallstone number, bile composition, and apolipoprotein E4 isoform. Hepatology. 1998;27:1508-1516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 31. | Ko CW, Beresford SA, Alderman B, Jarvik GP, Schulte SJ, Calhoun B, Tsuchida AM, Koepsell TD, Lee SP. Apolipoprotein E genotype and the risk of gallbladder disease in pregnancy. Hepatology. 2000;31:18-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 32. | Lin QY, Xiao LJ, Chen NS, Li N, Fu MD, Yan LN. A prospec-tive study on the serum lipids in different apo E genotype patients with gallstones. Zhonghua Yixue Yichuanxue Zazhi. 1997;14:223-226. |
| 33. | Ji Y, Wang N, Ramakrishnan R, Sehayek E, Huszar D, Breslow JL, Tall AR. Hepatic scavenger receptor BI promotes rapid clearance of high density lipoprotein free cholesterol and its transport into bile. J Biol Chem. 1999;274:33398-33402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 215] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 34. | Lammert F, Carey MC, Paigen B. Chromosomal organization of candidate genes involved in cholesterol gallstone formation: a murine gallstone map. Gastroenterology. 2001;120:221-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 101] [Article Influence: 4.2] [Reference Citation Analysis (0)] |