Copyright
©The Author(s) 2003.
World J Gastroenterol. Dec 15, 2003; 9(12): 2732-2736
Published online Dec 15, 2003. doi: 10.3748/wjg.v9.i12.2732
Published online Dec 15, 2003. doi: 10.3748/wjg.v9.i12.2732
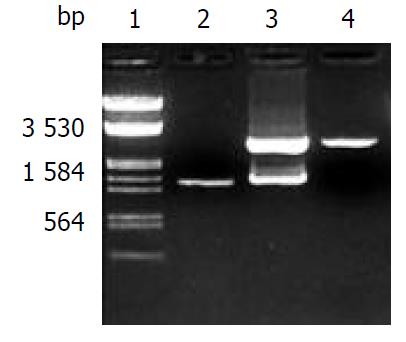
Figure 1 Electrophoretic identification of recombinant of pGEM-CYP2E1.
Lane 1: Markers (λ/EcoR I and Hind III), 2: PCR products of CYP2E1 (1.54 kb), 3: Recombinant of pGEM-CYP2E1 digested by Kpn I and Hind III, 4: pGEM-T vector.
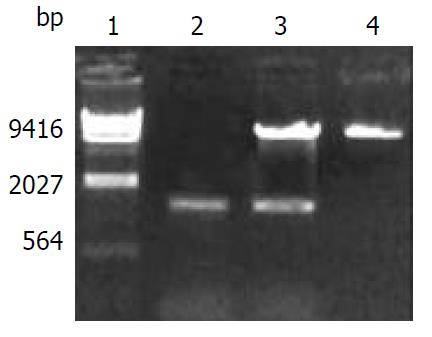
Figure 2 Electrophoretic identification of recombinant of pREP9-CYP2E1.
Lane 1: λDNA/Hind III Markers, 2: PCR prod-ucts of CYP2E1 (1.54 kb), 3: Recombinant of pREP9-CYP2E1 digested by Kpn I and Hind III, 4: pREP9 vector.
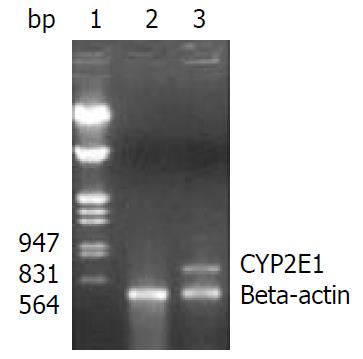
Figure 3 Identification of CYP2E1 mRNA expression in HepG2-CYP2E1 and HepG2 cells by RT-PCR with beta-actin as inter-nal control.
Lane 1: Markers (λ/EcoR I and Hind III), 2: RT-PCR products of HepG2 cells showing only beta-actin 462 bp, 3: RT-PCR products of HepG2-CYP2E1 cells showing 462 bp of beta-actin and 690 bp of CYP2E1.
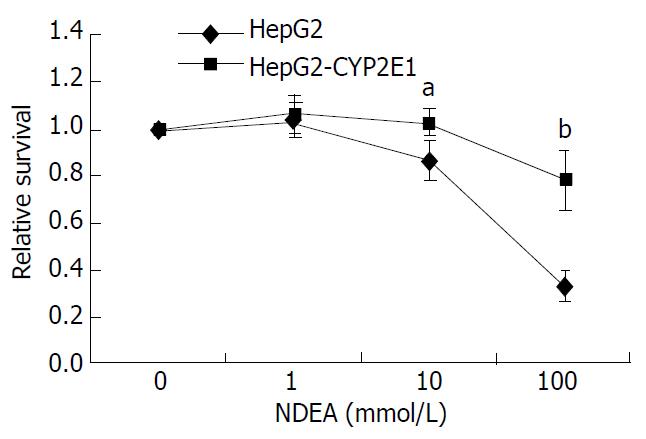
Figure 4 Cytotoxicity of NDEA against HepG2-CYP2E1 and HepG2 cells.
Cells were exposed to various concentrations of NDEA. Relative survival rate was represented as the relative toxicity to the control culture without NDEA. The results presented were the average of six duplications (-x±s). aP < 0.05, bP < 0.01 vs HepG2 cells.
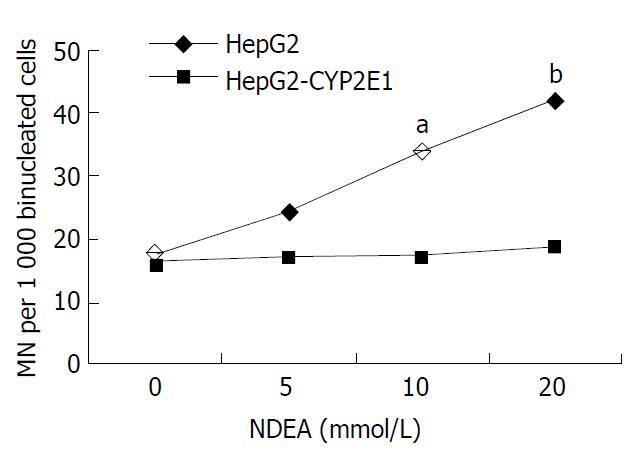
Figure 5 MN rates in HepG2-CYP2E1 and HepG2 cells in-duced by NDEA.
Cells were exposed to various concentrations of NDEA. The data were expressed as per thousand of binucleated cells with MN. aP < 0.05, bP < 0.01 vs HepG2 cells.
- Citation: Zhuge J, Luo Y, Yu YN. Heterologous expression of human cytochrome P450 2E1 in HepG2 cell line. World J Gastroenterol 2003; 9(12): 2732-2736
- URL: https://www.wjgnet.com/1007-9327/full/v9/i12/2732.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i12.2732