Copyright
©The Author(s) 2025.
World J Gastroenterol. Apr 28, 2025; 31(16): 104920
Published online Apr 28, 2025. doi: 10.3748/wjg.v31.i16.104920
Published online Apr 28, 2025. doi: 10.3748/wjg.v31.i16.104920
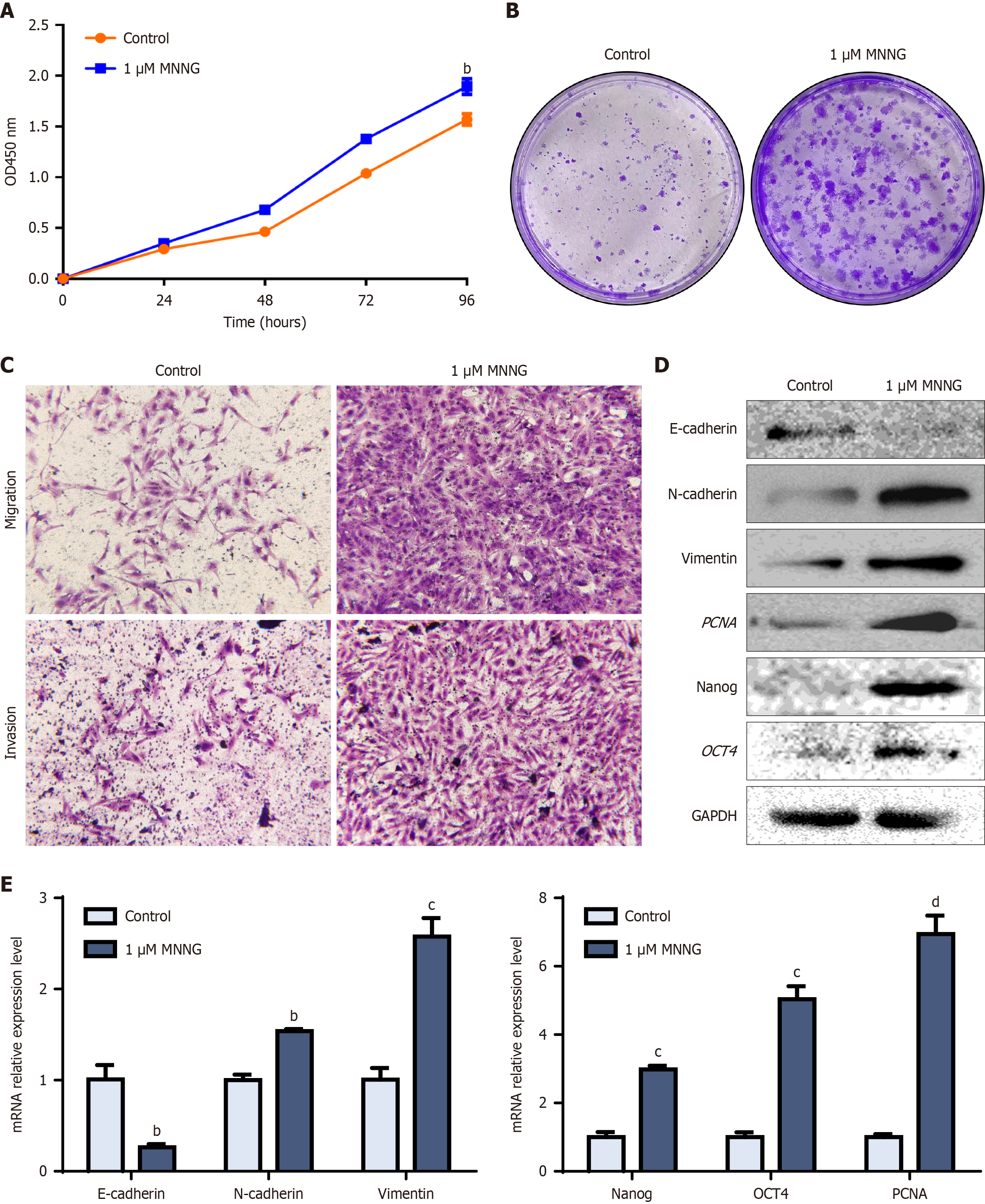
Figure 1 N-methyl-N’-nitro-N-nitrosoguanidine-induced malignant transformation of GES-1 cells.
A: Cell Counting Kit-8 detection of the proliferative capacity of GES-1 cells following chronic N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) exposure across 30 generations; B: Changes in the clonogenic capacity of MNNG-treated GES-1 cells; C: Alterations in the migratory invasive capacities of MNNG-treated GES-1 cells; D: Protein levels of epithelial-to-mesenchymal transition (EMT), stemness, and proliferative markers; E: mRNA levels of EMT, stemness, and proliferative markers. bP < 0.01. cP < 0.001. dP < 0.0001. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; OD: Optical density.
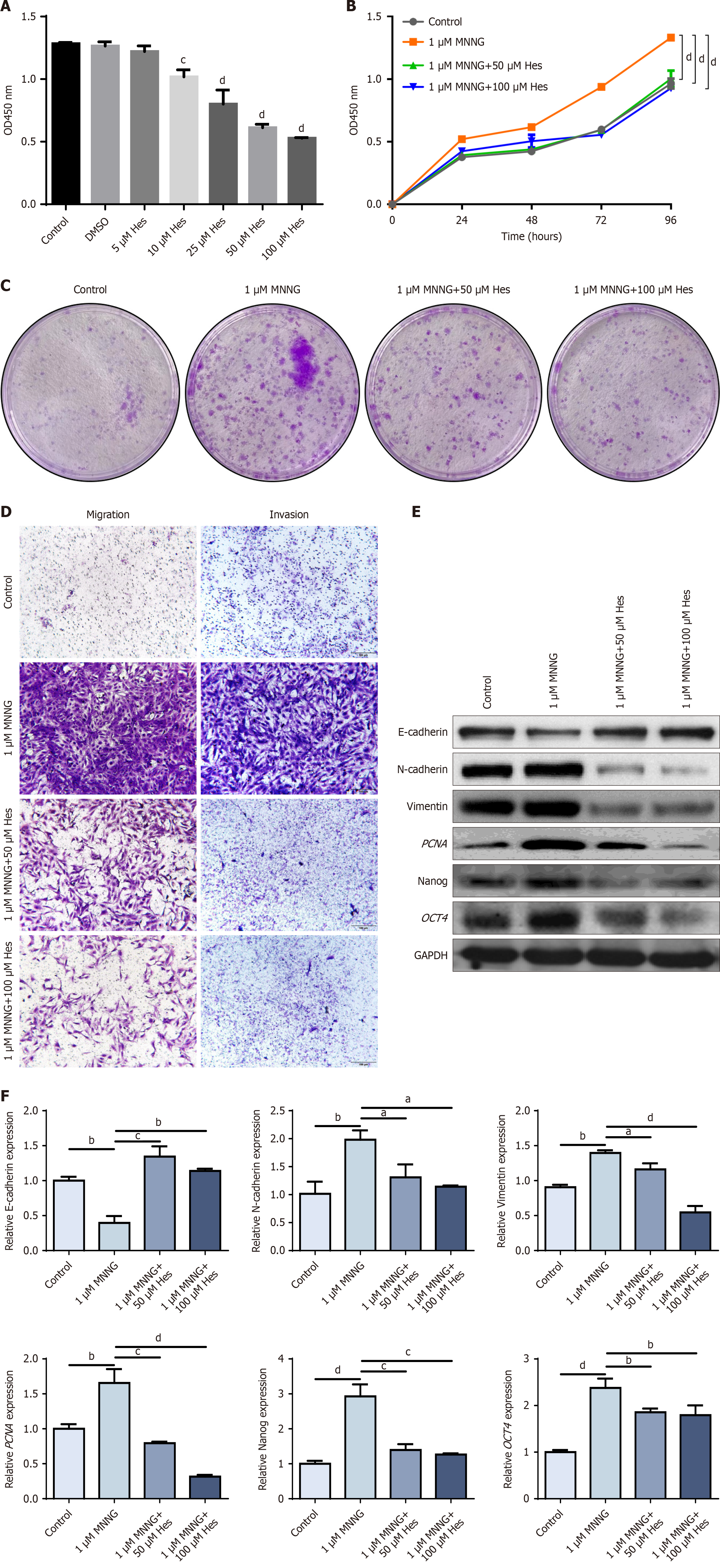
Figure 2 Intervention effect of hesperetin on TGES-1 cells.
A: Cell Counting Kit-8 (CCK-8) detection of the survival rate of TGES-1 cells; B: CCK-8 detection of the proliferation ability of TGES-1 cells; C: Alteration of the clonogenic ability of TGES-1 cells; D: Alteration of the migratory and invasive abilities of TGES-1 cells; E: Protein levels of epithelial-to-mesenchymal transition (EMT), stemness, and proliferative markers; F: mRNA levels of EMT, stemness, and proliferative markers. aP < 0.05. bP < 0.01. cP < 0.001. dP < 0.0001. DMSO: Dimethyl sulfoxide; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; MNNG: N-methyl-N’-nitro-N-nitrosoguanidine; OD: Optical density.
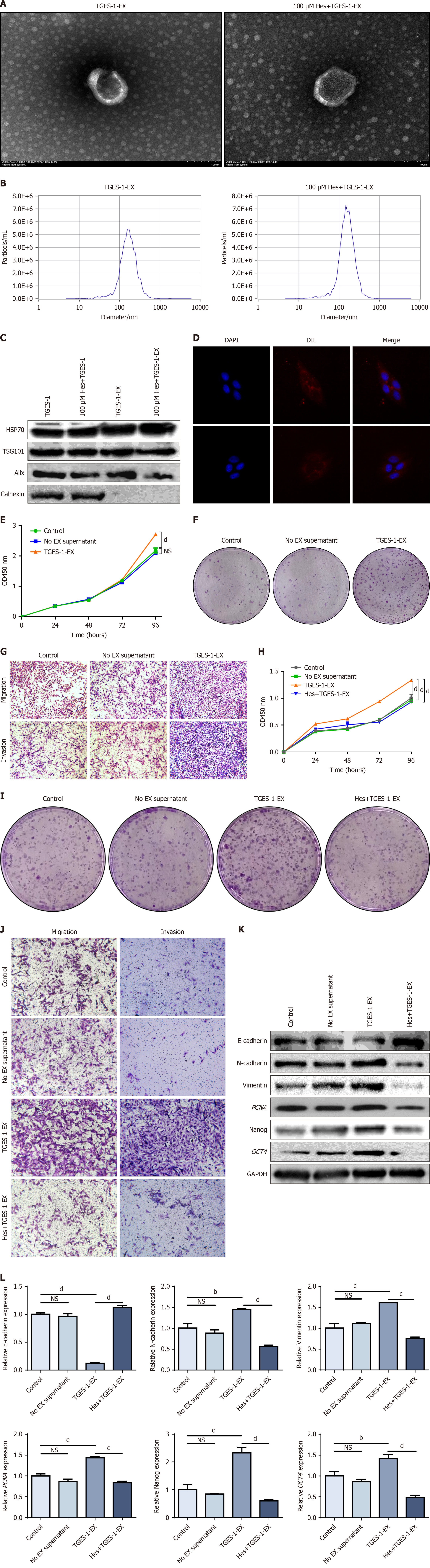
Figure 3 Exosomes derived from TGES-1 cells promoted GES-1 cell proliferation, migration, and invasion.
A: Transmission electron microscopy detected exosome morphology; B: Nanoparticle tracking analysis detected exosome particle size; C: Exosomal surface protein expression was characterized by Western blotting; D: 4’,6-diamidino-2-phenylindole blue fluorescence labeling of nuclei, Dil red fluorescence labeling of exosomes, and confocal microscopy showed that exosomes were taken up by GES-1 cells; E: Cell Counting Kit-8 detection of proliferative capacity of exosomes derived from TGES-1 cells (TGES-1-EX) after action on GES-1 cells; F: Clonogenic capacity of TGES-1-EX after action on GES-1 cells; G: Migration invasion ability of TGES-1-EX after action on GES-1 cells; H: Proliferative capacity after hesperetin-pretreated TGES-1-EX action on GES-1 cells; I: Clonogenic capacity of TGES-1 cells pretreated with hesperetin after action on GES-1 cells; J: Invasion and migration ability of GES-1 cells after treatment with exosomes; K: Protein levels of epithelial-to-mesenchymal transition (EMT), stemness, and proliferative markers; L: mRNA expression of EMT, stemness, and proliferative markers. aP < 0.05. bP < 0.01. cP < 0.001. dP < 0.0001. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; MNNG: N-methyl-N’-nitro-N-nitrosoguanidine; OD: Optical density.
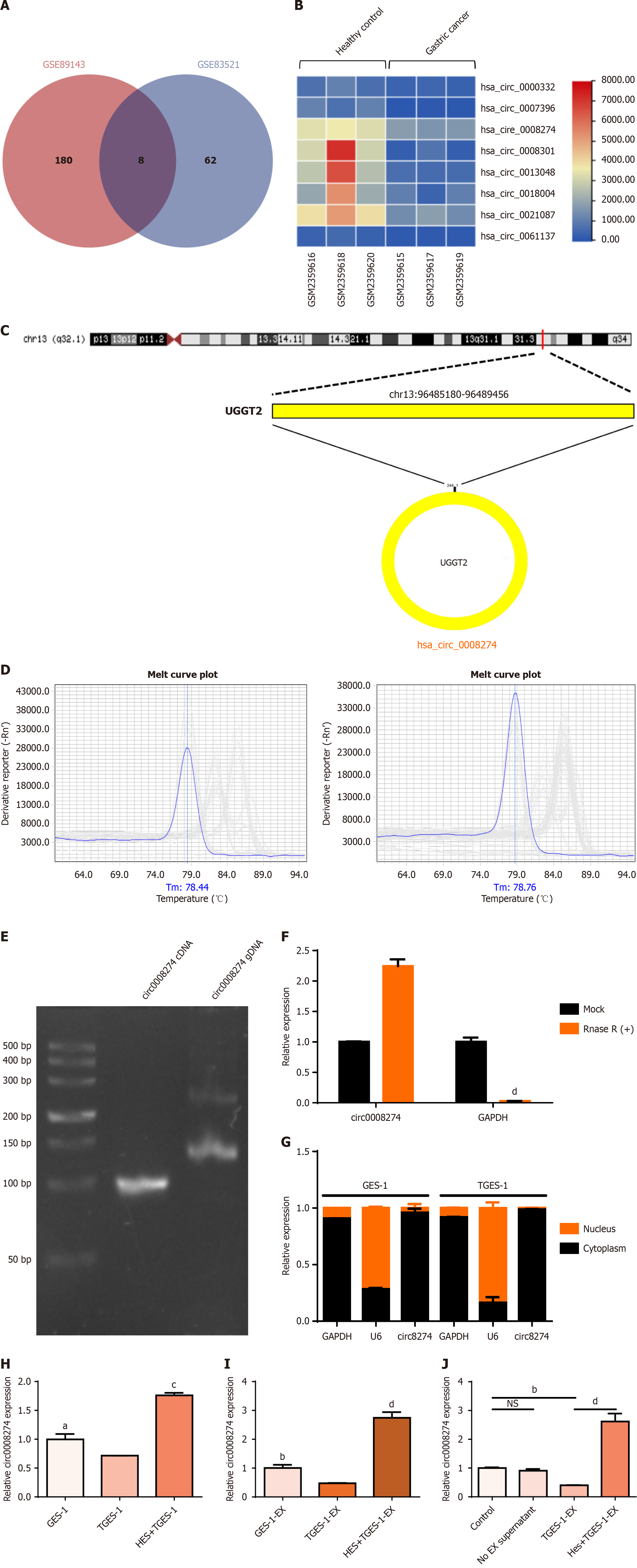
Figure 4 Identification of circ0008274 and its expression in cells and exosomes.
A: Differently expressed circular RNAs (circRNAs) overlapping in public databases; B: Hierarchical clustering analysis of differentially expressed circRNAs as visualized in the heat map; C: Circ0008274 cyclisation mechanism; D: Cellular RNA and exosomes RNA quantitative polymerase chain reaction (qPCR) melting curves; E: Agarose gel electrophoresis; F: qPCR to detect before and after treatment with RNase R circ0008274 expression level; G: Nucleoplasmic separation to determine circ0008274 localization; H: Effect of hesperetin on the level of circ0008274 in cells; I: Modulatory effects of hesperetin on circ0008274 expression in exosomes; J: Expression of circ0008274 in exosomes derived from TGES-1 cells and hesperetin-pretreated TGES-1 cell exosomes after action on GES-1 cells. aP < 0.05. bP < 0.01. cP < 0.001. dP < 0.0001. cDNA: Complementary DNA; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; gDNA: Genomic DNA; TGES-1-EX: Exosomes derived from TGES-1 cells.
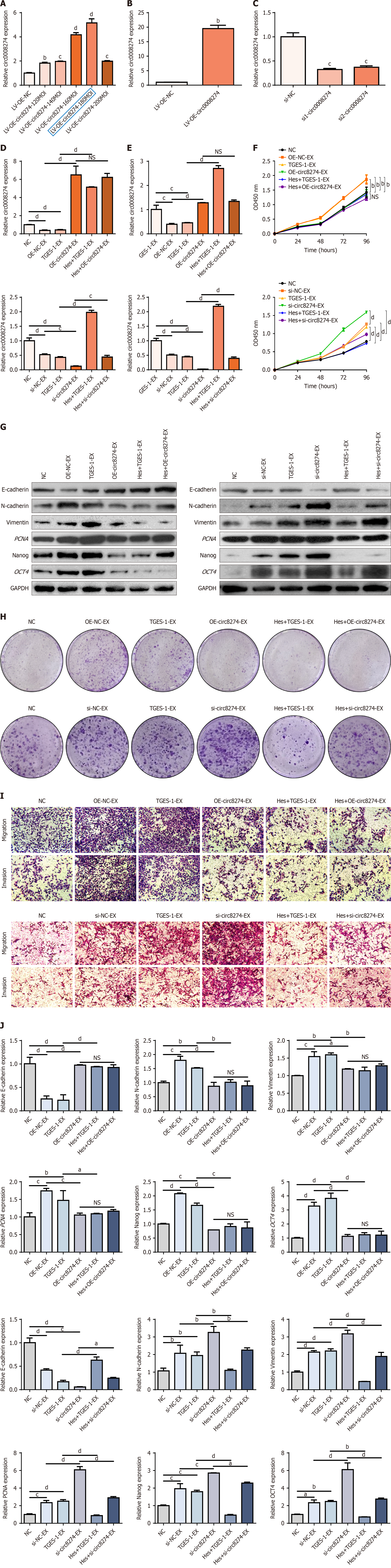
Figure 5 Hesperetin pretreatment of exosomes from overexpressed/knockdown circ0008274 TGES-1 cells action on GES-1 cells.
A: Expression level of circ0008274 after transfection with overexpressing lentivirus; B: Level of circ0008274 in TGES-1 cells after puromycin screening; C: Expression of circ0008274 after TGES-1 transfection with small interfering RNA; D: Circ0008274 expression in exosomes of hesperetin pretreated overexpressing/knockdown circ0008274 TGES-1 cells; E: Circ0008274 expression after hesperetin pretreatment overexpression/knockdown of exosomes of circ0008274 TGES-1 cells acting on GES-1 cells; F: Changes in growth capacity of GES-1 cells treated with exosomes from different groups; G: Protein levels of epithelial-to-mesenchymal transition (EMT), stemness and proliferation markers in GES-1 cells treated with hesperetin and overexpression/knockdown circ0008274 exosomes derived from TGES-1 cells (TGES-1-EX); H: Changes in clonogenic capacity of GES-1 cells treated with exosomes from different groups; I: Changes in migration and invasion capacity of GES-1 cells treated with exosomes from different groups; J: mRNA expression of EMT, stemness and proliferation markers in GES-1 cells treated with hesperetin and overexpression/knockdown circ0008274 TGES-1-EX. aP < 0.05. bP < 0.01. cP < 0.001. dP < 0.0001. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; LV: Lentivirus vector; NC: Negative control; OD: Optical density; OE: Overexpression; si-NC: Small interfering RNA negative control; si-NC-EX: Small interfering RNA negative control-exosome; TGES-1-EX: Exosomes derived from TGES-1 cells.
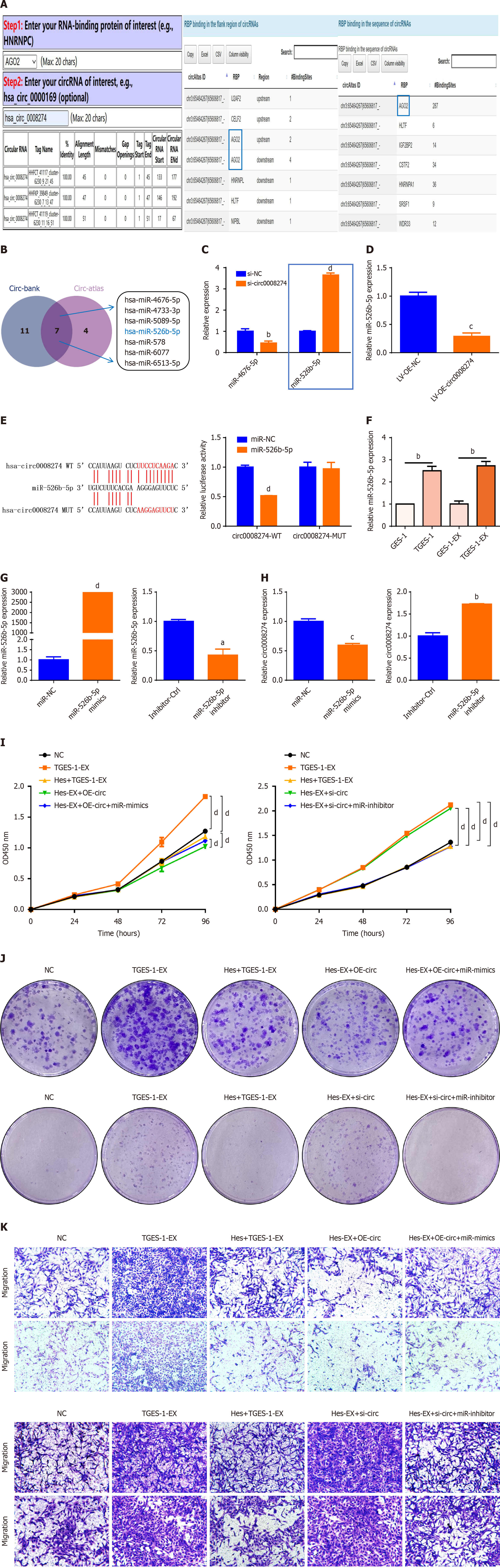
Figure 6 Hesperetin mediated N-methyl-N’-nitro-N-nitrosoguanidine induced gastric cancer by targeting miR-526b-5p through exosomal circ0008274.
A: Circlnteractome and circ-atlas databases showed that Argonaute 2 has the ability to bind to circ0008274; B: Circ-bank and circ-atlas databases predicted the microRNAs (miRNAs) that circ0008274 binds to and take the intersections; C: MiR-526b-5p was highly expressed and miR-4676-5p was lowly expressed after circ0008274 knockdown; D: MiR-526b-5p was lowly expressed after overexpression of circ0008274; E: Dual-luciferase reporter gene assay; F: Quantitative polymerase chain reaction detection of miR-526b-5p expression in cells and exosomes; G: MiR-526b-5p expression after transfection of miR-526b-5p mimics/inhibitor; H: Circ0008274 expression after transfection of miR-526b-5p mimics/inhibitor; I: Proliferative capacity of TGES-1 cells after treated with exosomes from different groups; J: Changes in clonogenic capacity of TGES-1 cells treated with exosomes from different groups; K: Changes in migration and invasion capacity of TGES-1 cells treated with exosomes from different groups. aP < 0.05. bP < 0.01. cP < 0.001. dP < 0.0001. EX: Exosome; LV: Lentivirus vector; MUT: Mutant; NC: Negative control; OD: Optical density; OE: Overexpression; si-NC: Small interfering RNA negative control; TGES-1-EX: Exosomes derived from TGES-1 cells; WT: Wild-type.
- Citation: Liang ZF, Xu YM, Song JJ, Gao ZH, Qian H, Xu XZ. Interventional effect of hesperetin on N-methyl-N’-nitro-N-nitrosoguanidine-induced exosomal circ008274 in affecting normal cells to promote gastric carcinogenesis. World J Gastroenterol 2025; 31(16): 104920
- URL: https://www.wjgnet.com/1007-9327/full/v31/i16/104920.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i16.104920