Copyright
©The Author(s) 2024.
World J Gastroenterol. Jul 21, 2024; 30(27): 3290-3303
Published online Jul 21, 2024. doi: 10.3748/wjg.v30.i27.3290
Published online Jul 21, 2024. doi: 10.3748/wjg.v30.i27.3290
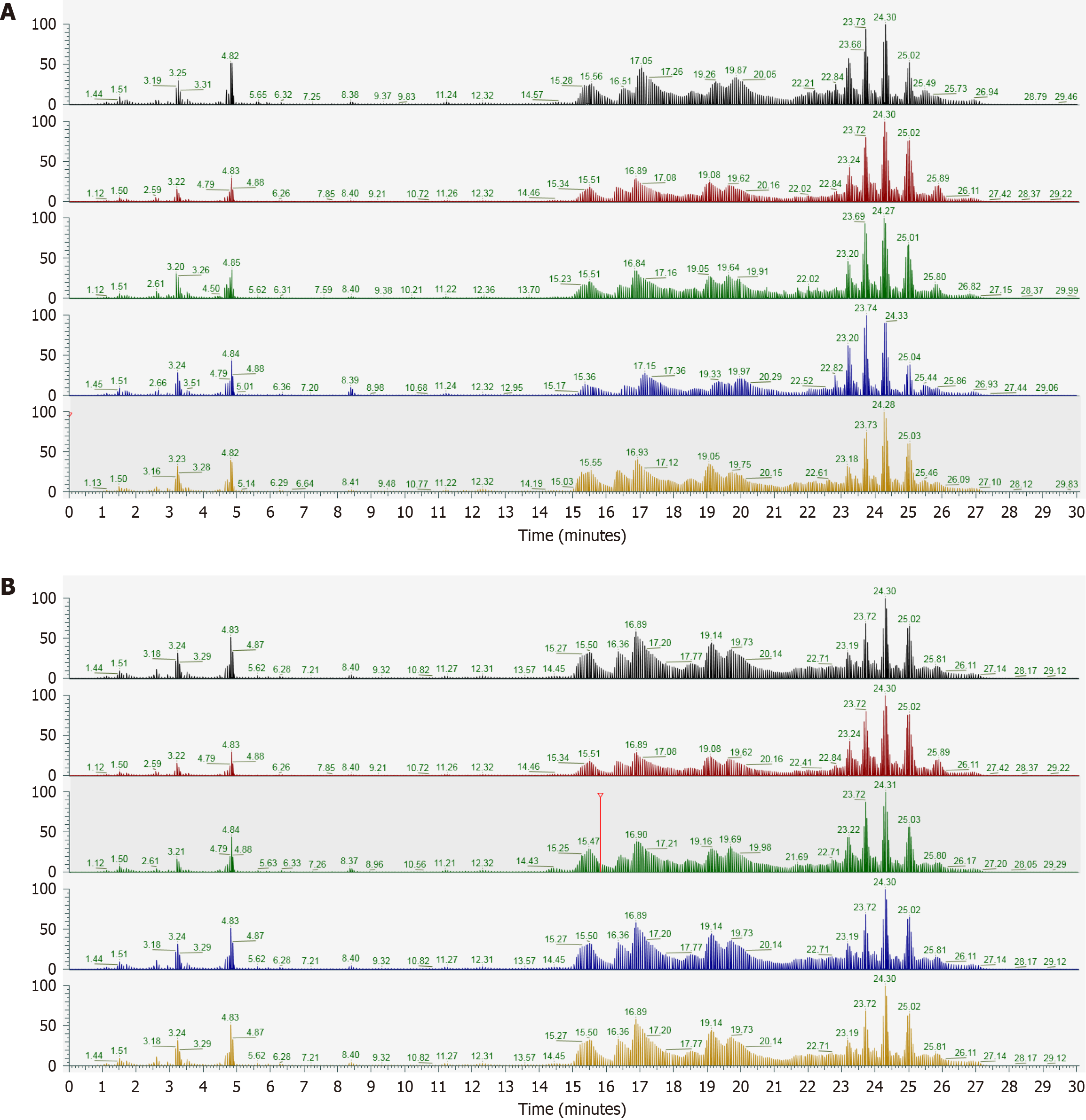
Figure 1 Total ion chromatogram.
A: Negative mode; B: Positive mode.
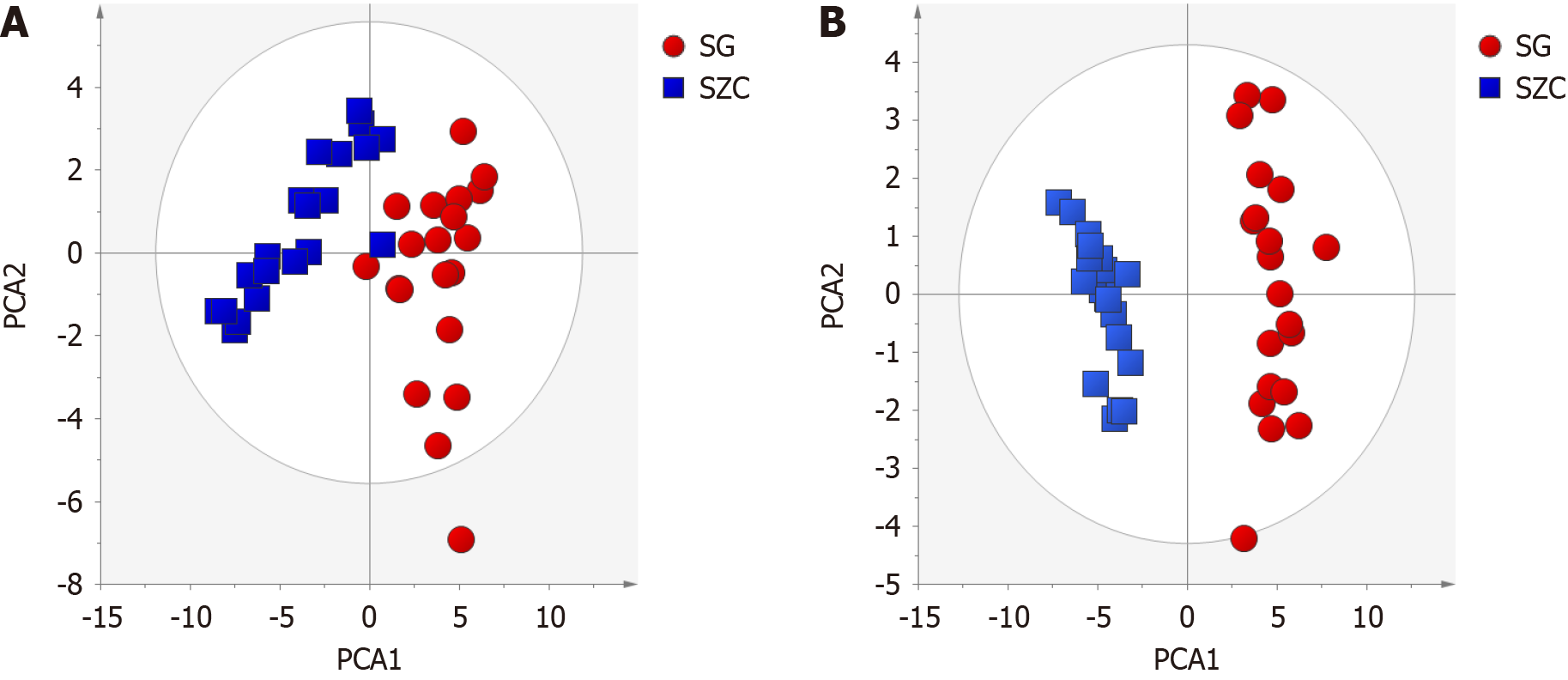
Figure 2 Principal component analysis.
A: Positive mode; B: Negative mode. SG: Lean metabolic-associated fatty liver disease (metabolic-associated fatty liver disease); SZC: Healthy lean individuals.
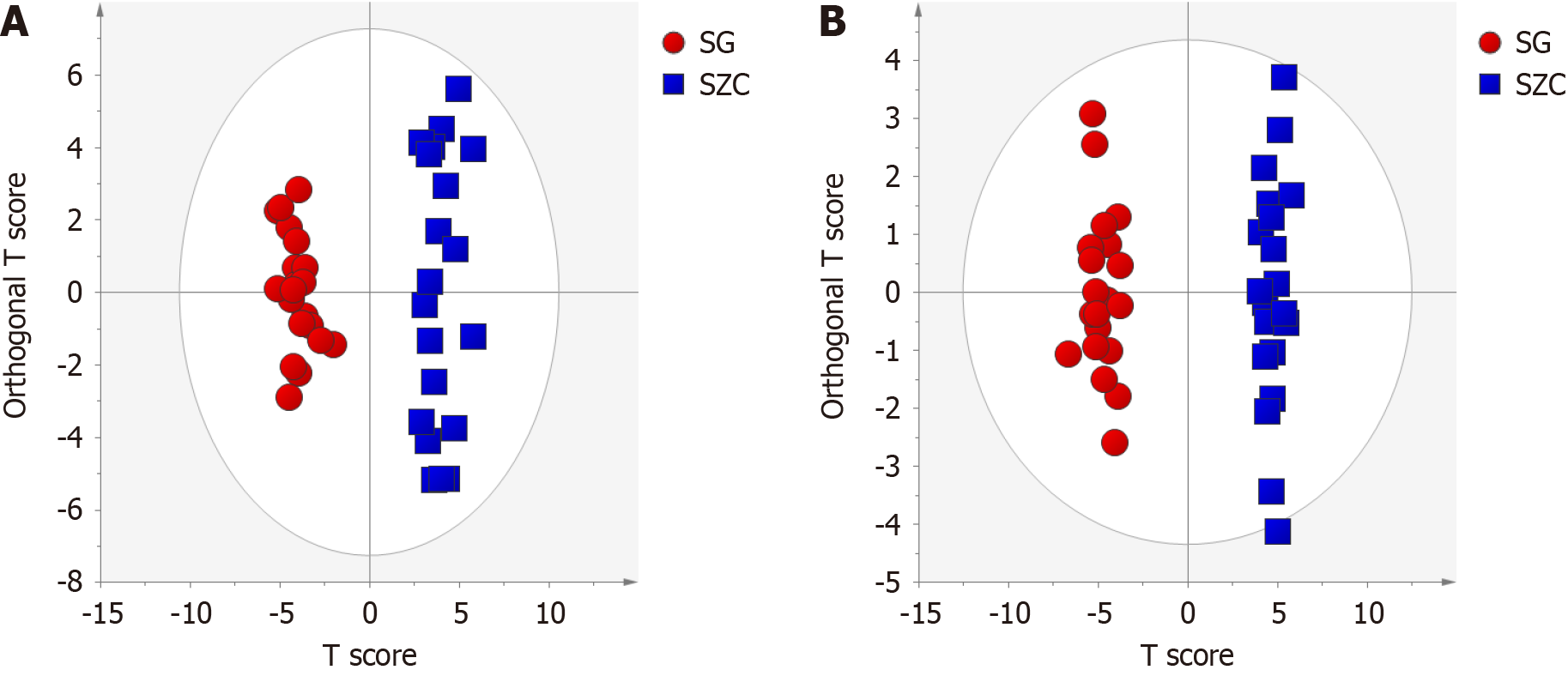
Figure 3 Orthogonal partial least squares discriminant analysis.
A: Positive mode; B: Negative mode. SG: Lean-type metabolic-associated fatty liver disease (metabolic-associated fatty liver disease); SZC: Lean healthy individuals.
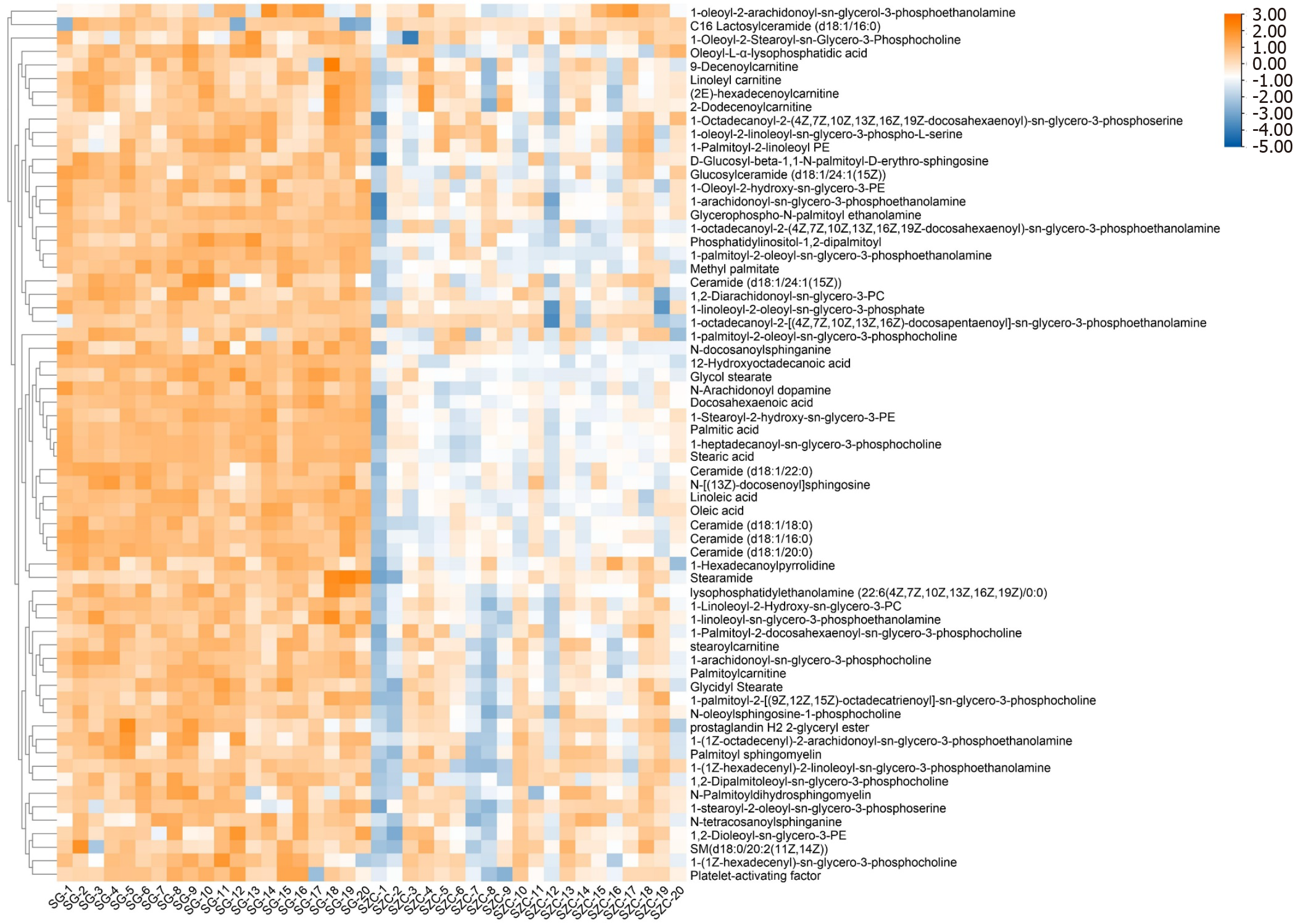
Figure 4 Heatmap of differential metabolites.
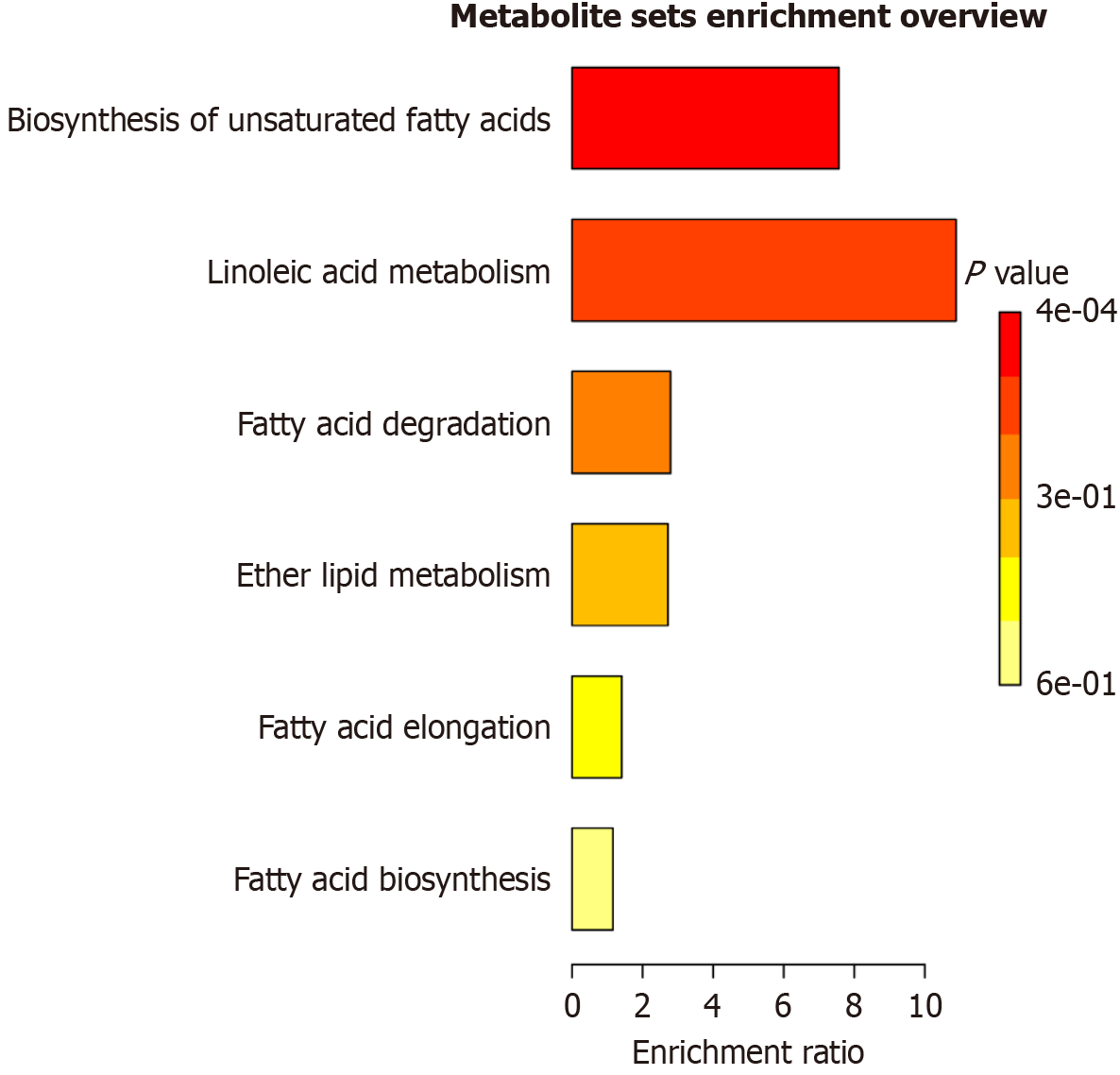
Figure 5 Metabolic pathway enrichment analysis.
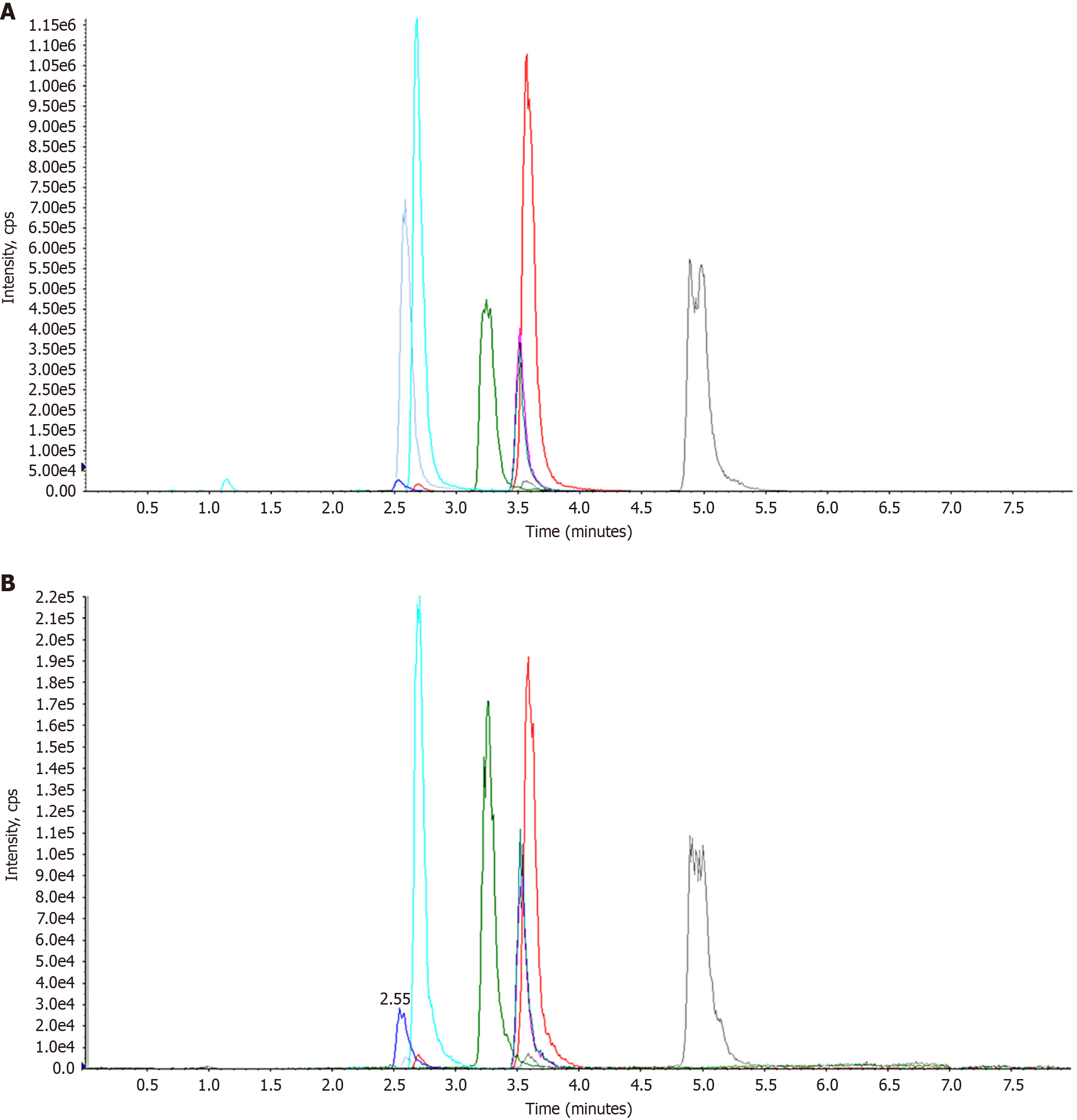
Figure 6 Standard material and sample total ion chromatograms.
A: Standard material; B: Sample total.
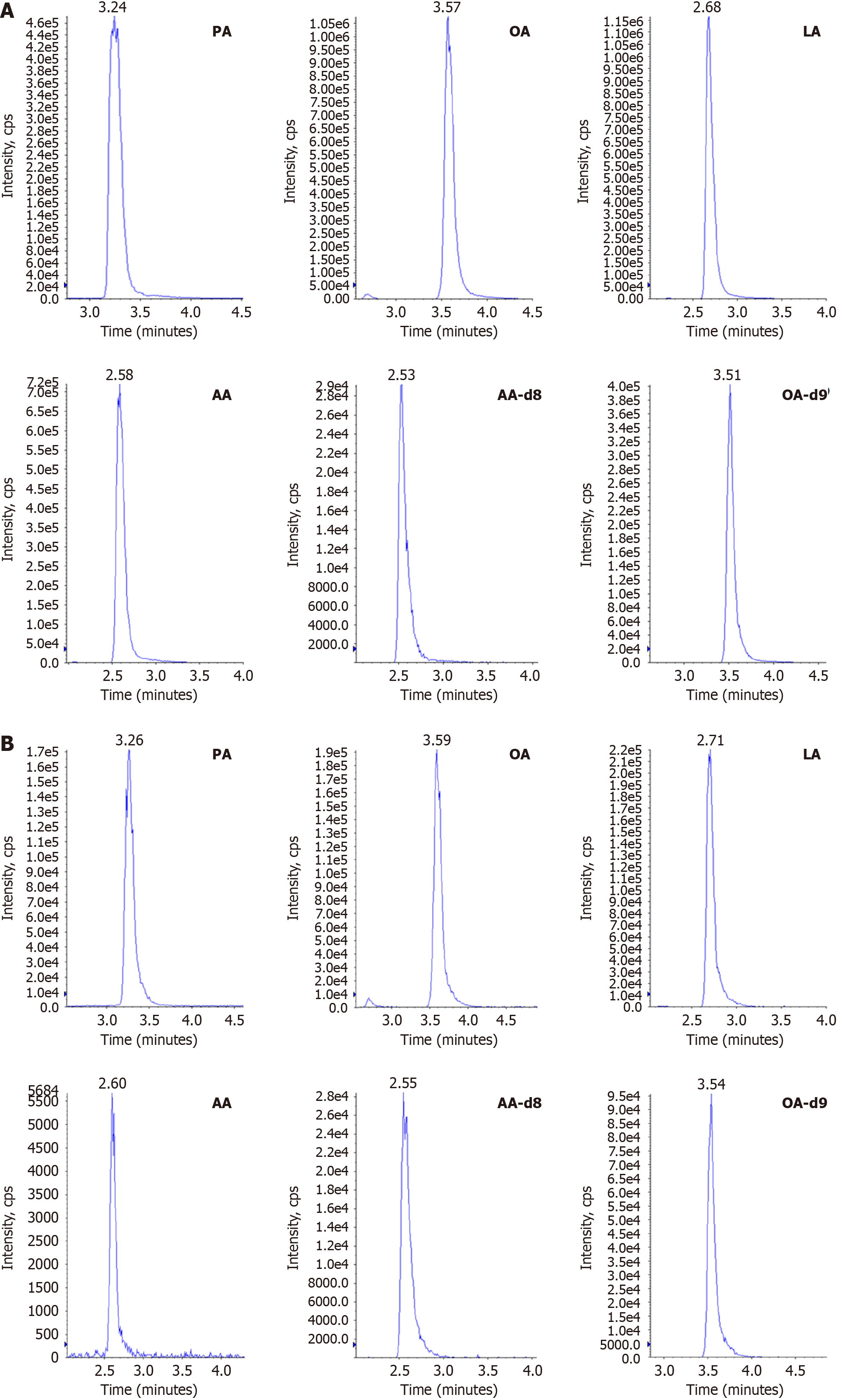
Figure 7 Multiple reaction monitoring chromatograms of palmitic acid, oleic acid, linoleic acid, arachidonic acid, and internal standards.
A: Standards; B: Samples. PA: Palmitic acid; OA: Oleic acid; LA: Linoleic acid; AA: Arachidonic acid; OA-d9: Deuterated oleic acid.
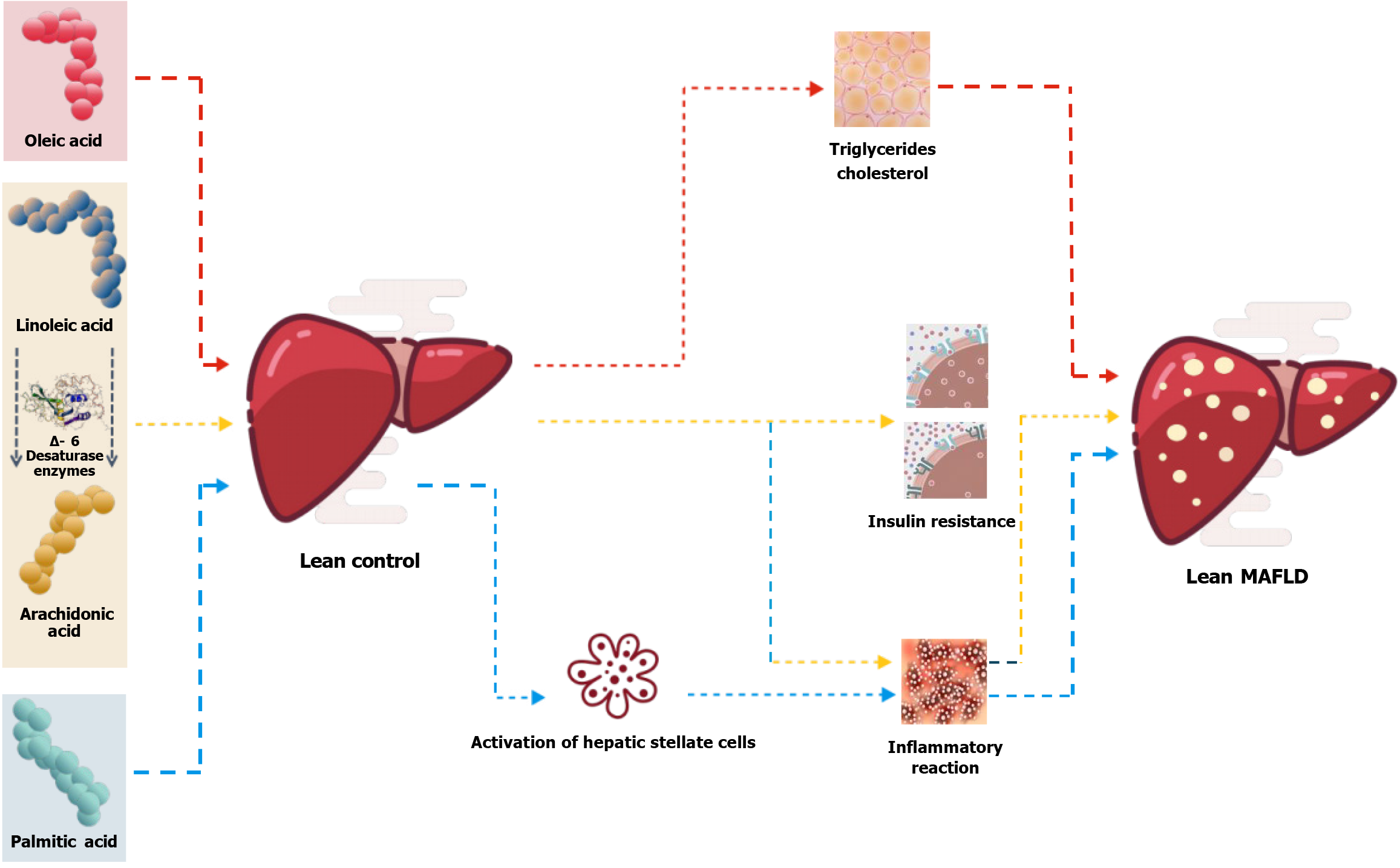
Figure 8 Effects of fatty acids on metabolic-associated fatty liver disease.
Through serum targeted metabolomics studies, we measured four specific biomarkers. The preliminary conclusion was that the transformation of lean control individuals to lean metabolic-associated fatty liver disease (MAFLD) may affect the Oleic acid, linoleic-arachidonic acid and palmitic acid pathways. Linoleic acid is converted to arachidonic acid by Δ-6 desaturase, as shown in the figure.
- Citation: Sun PQ, Dong WM, Yuan YF, Cao Q, Chen XY, Guo LL, Jiang YY. Targeted metabolomics study of fatty-acid metabolism in lean metabolic-associated fatty liver disease patients. World J Gastroenterol 2024; 30(27): 3290-3303
- URL: https://www.wjgnet.com/1007-9327/full/v30/i27/3290.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i27.3290