Copyright
©The Author(s) 2022.
World J Gastroenterol. Jul 7, 2022; 28(25): 2782-2801
Published online Jul 7, 2022. doi: 10.3748/wjg.v28.i25.2782
Published online Jul 7, 2022. doi: 10.3748/wjg.v28.i25.2782
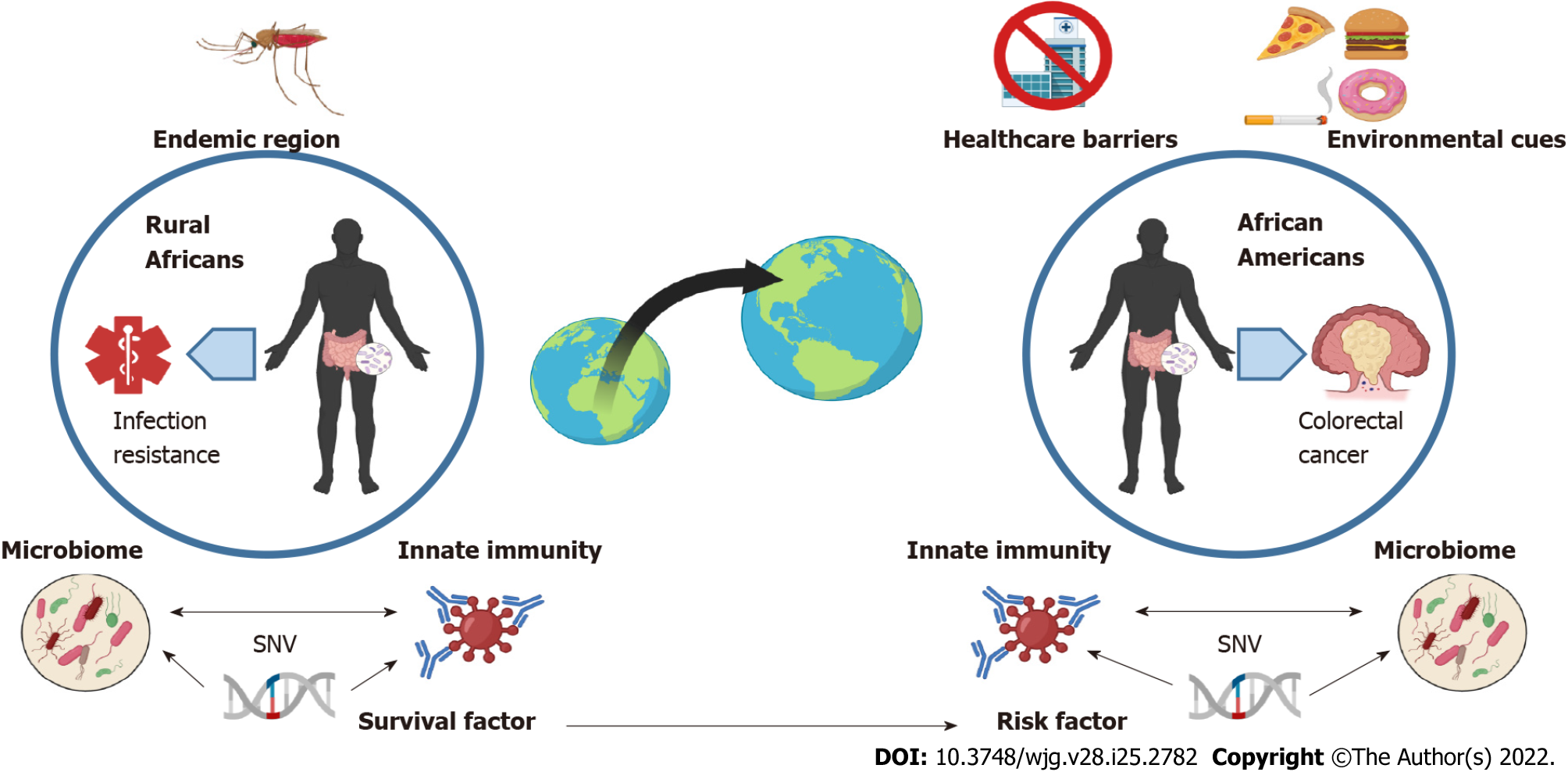
Figure 1 Immune-related variant may promote survival to pathogens in ancestral African environment but precipitate cancer in descendent African-Americans.
Pathogens associated with endemic African regions (e.g., malaria) are thought to pressure selection for specific immune-related genetic variants associated with pathogen resistance and survival of Native Africans (left). In the context of westernized diet and lifestyle, this genetic predisposition (represented herein by a single nucleotide variant), when associated with inflammatory regulation and inherited from African ancestors, may lead to altered interactions with bacteria or communities of bacteria of the gut microbiome, thereby precipitating the colon adenoma-carcinoma sequence in African-Americans (right). Higher inflammation associated with lack of exercise, high fat diet, and socio-economic status are thought to be predominant factors driving early colorectal cancer onset in African-Americans via their impact on shaping the gut microbiome and its interactions with the host genetic background. SNV: Single nucleotide variant. Created with Biorender.com.
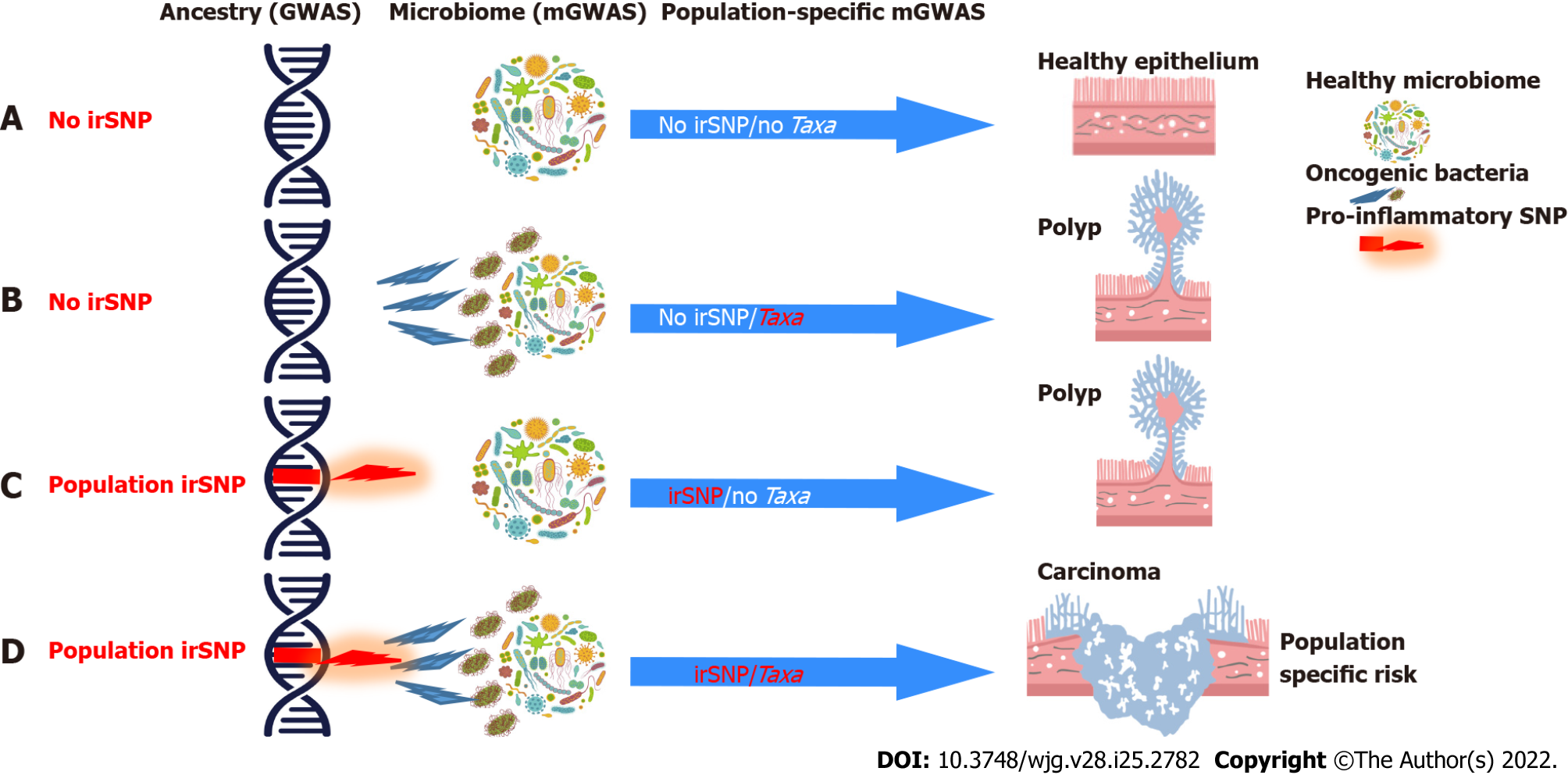
Figure 2 Interactions between host genetics and microbiome delineate colorectal cancer risk factors.
Immune-related risk factors of colorectal cancer (CRC) may combine host genetics [including immune-related single-nucleotide polymorphisms (irSNPs)] and microbiome cues. The genetic regulation (irSNP) of bacteria (Taxa) recognition by the mucosal immunity may mediate the heterogeneity of CRC risk factors between populations which vary by the representation of irSNPs. A: Absence of irSNPs and pathogenic bacteria (no irSNPs/no Taxa) maintain healthy mucosal inflammation and gut barrier; B: Absence of irSNPs with enrichment of pathogenic bacteria (no irSNPs/Taxa) may lead to chronic inflammation that slowly promotes polyp formation; C: Presence of irSNP in absence of pathogenic bacteria may also lead to chronic inflammation that slowly promotes polyp formation; D: Combination of irSNP and pathogenic bacteria (dysbiosis) may trigger a smoldering chronic inflammatory response precipitating early progression of the adenoma-carcinoma sequence. Genome-wide association studies (GWAS) (irSNP as CRC risk) and microbiome GWAS (mGWAS) (bacteria as CRC risk) are poised to miss the association of the irSNP with CRC because of the necessity of a combined occurrence of irSNP and specific bacteria enrichment in the microbiota to increase the CRC risk. However, if the irSNP is linked to the genetic ancestry, population-specific microbiome GWAS will likely detect the association of irSNP and the bacteria as a CRC risk in the population carrying this genetic ancestry. CRC: Colorectal cancer; irSNPs: Immune-related single-nucleotide polymorphisms; GWAS: Genome-wide association studies; mGWAS: Microbiome genome-wide association studies.
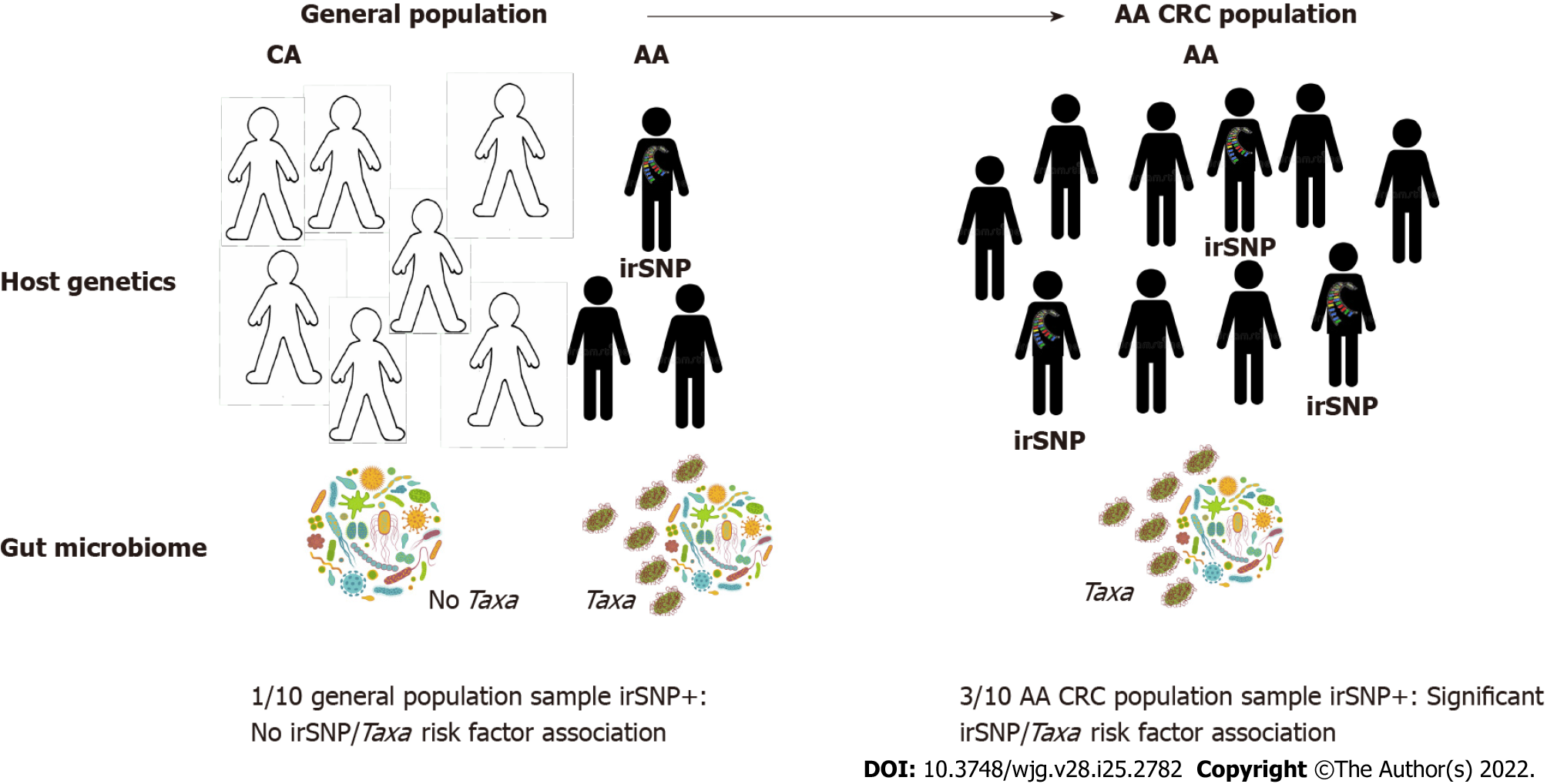
Figure 3 Population-specific colorectal cancer microbiome genome-wide association studies to understand colorectal cancer disparities.
Systemic underrepresentation of minorities in microbiome genome-wide association studies (mGWAS) may compromise the identification of microbiome-associated colorectal cancer (CRC) risks in African-Americans (AAs). Left: Associations between AA-enriched immune-related single-nucleotide polymorphisms (irSNPs) (pop-irSNPs) and gut bacteria (Taxa) may remain undetected in the global population if pop-irSNP and Taxa have too low frequency in the AA population and AAs are underrepresented. Right: If pop-irSNP and pathogenic bacteria are CRC risk factors in AA, they will be enriched in AA CRC patients compared to the general AA population (left), and mGWAS will detect the association between pop-irSNP and Taxa in AA CRC cohort. These features justify population-specific CRC mGWAS to detect additional CRC risk resulting from the interaction between ancestral irSNP and bacteria in minorities. CRC: Colorectal cancer; irSNPs: Immune-related single-nucleotide polymorphisms; mGWAS: Microbiome genome-wide association studies; AA: African-American; CA: Caucasian-American.
- Citation: Ahmad S, Ashktorab H, Brim H, Housseau F. Inflammation, microbiome and colorectal cancer disparity in African-Americans: Are there bugs in the genetics? World J Gastroenterol 2022; 28(25): 2782-2801
- URL: https://www.wjgnet.com/1007-9327/full/v28/i25/2782.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i25.2782