Copyright
©The Author(s) 2021.
World J Gastroenterol. Jul 28, 2021; 27(28): 4555-4581
Published online Jul 28, 2021. doi: 10.3748/wjg.v27.i28.4555
Published online Jul 28, 2021. doi: 10.3748/wjg.v27.i28.4555
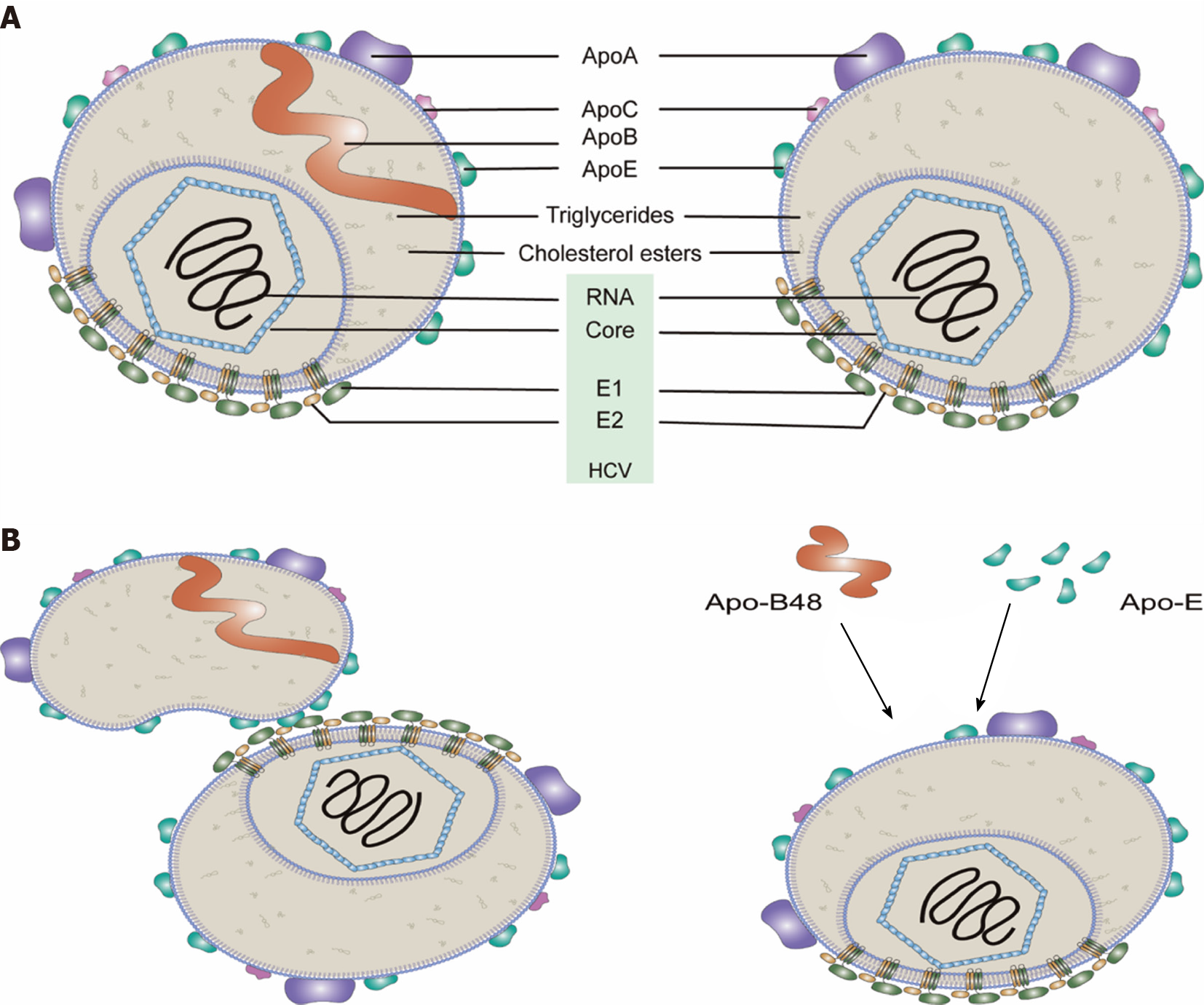
Figure 1 Model of hepatitis C virus particles (lipo-viral particles) secreted from cells.
A: Most of the lipo-viral particles (LPV)’s membrane is a lipid monolayer. A bilayer-containing region is where the viral envelope proteins (i.e., E1 and E2) are inserted. Viral envelope proteins may also exist in the phospholipid monolayer membrane. Though the precise structure has not yet been determined, LVPs are believed to have multiple copies of Apo-E and less Apo-A1 molecules but only one Apo-B100 molecule (left). Some LPVs may not have Apo-B (right). Within the phospholipid monolayer, there are the core proteins wrapping the viral RNA genome and neutral lipids (e.g., cholesterol esters and triglycerides); B: Serum lipoproteins are possibly associated with hepatitis C virus particles in different ways. HCV: Hepatitis C virus.
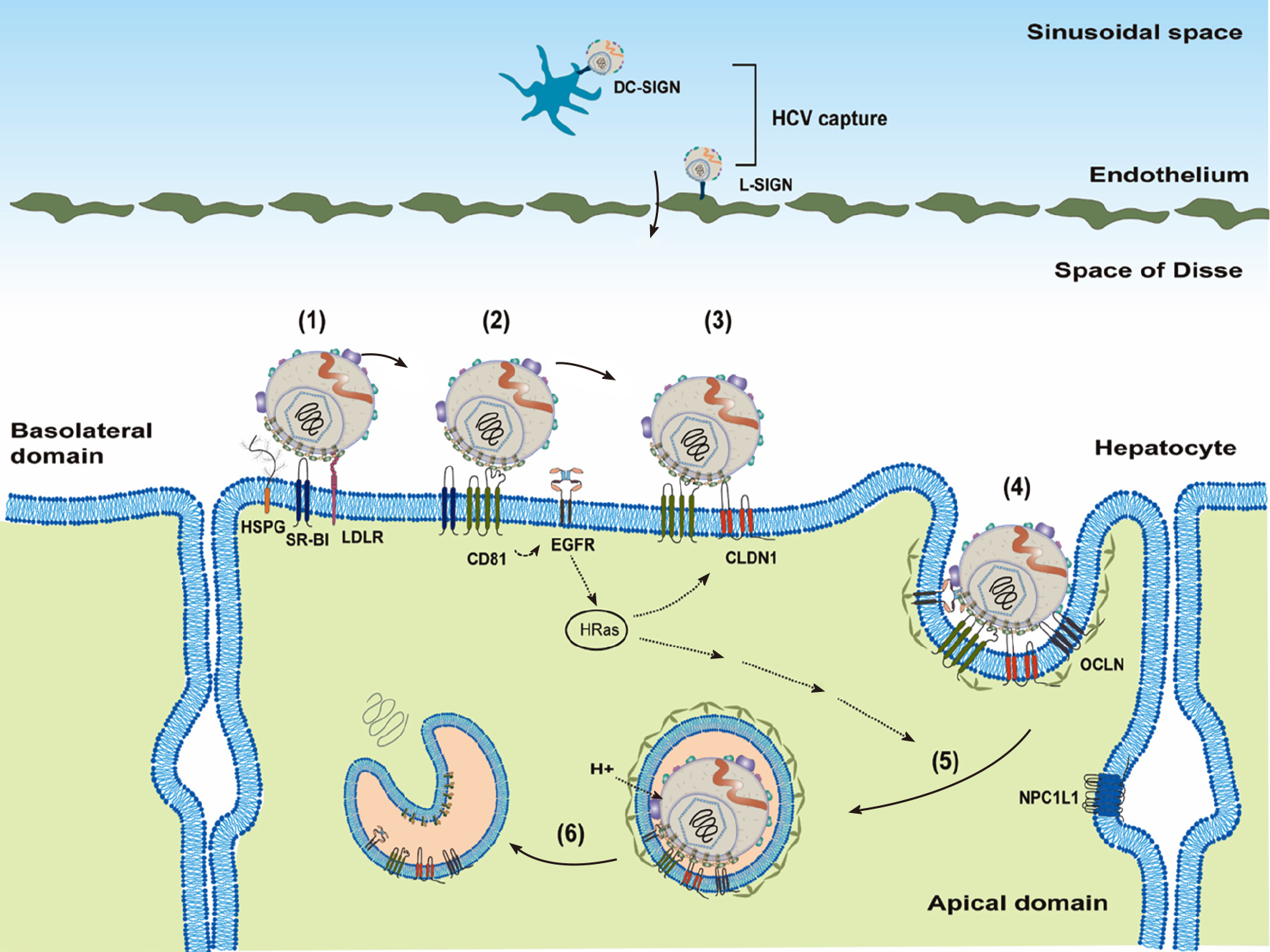
Figure 2 Model of cell-free virus entry into hepatocytes.
Hepatitis C virus (HCV) lipo-viral particles (LPVs) may be captured by DC-SIGN on the dendritic cells or L-SIGN on the endothelium in the sinusoidal space. After transfer to Space of Disse, HCV LPVs could attach to the hepatocytes through interacting with highly sulfated heparan sulfate proteoglycans, low-density lipoprotein receptor and scavenger receptor class B type 1 (1). This attachment allows the engagement of LPVs to cluster of differentiation 81 (CD81) and then induces the epidermal growth factor receptor receptor signaling (2). Lateral diffusion of the CD81–HCV complexes results in the association of CD81–HCV with Claudin-1 (3) and then OCLIN (4). Formation of the HCV–CD81–CLDN1–OCLIN complex allows viral particles internalized through clathrin-dependent endocytosis (5). Endosomal acidification induces the fusion of viral particles possibly through E1 and leads to the release of the viral genomic RNA into cytosol (6). HSPGs: Heparan sulfate proteoglycans; HCV: Hepatitis C virus; EGFR: Epidermal growth factor receptor; LDLR: Low-density lipoprotein receptor.
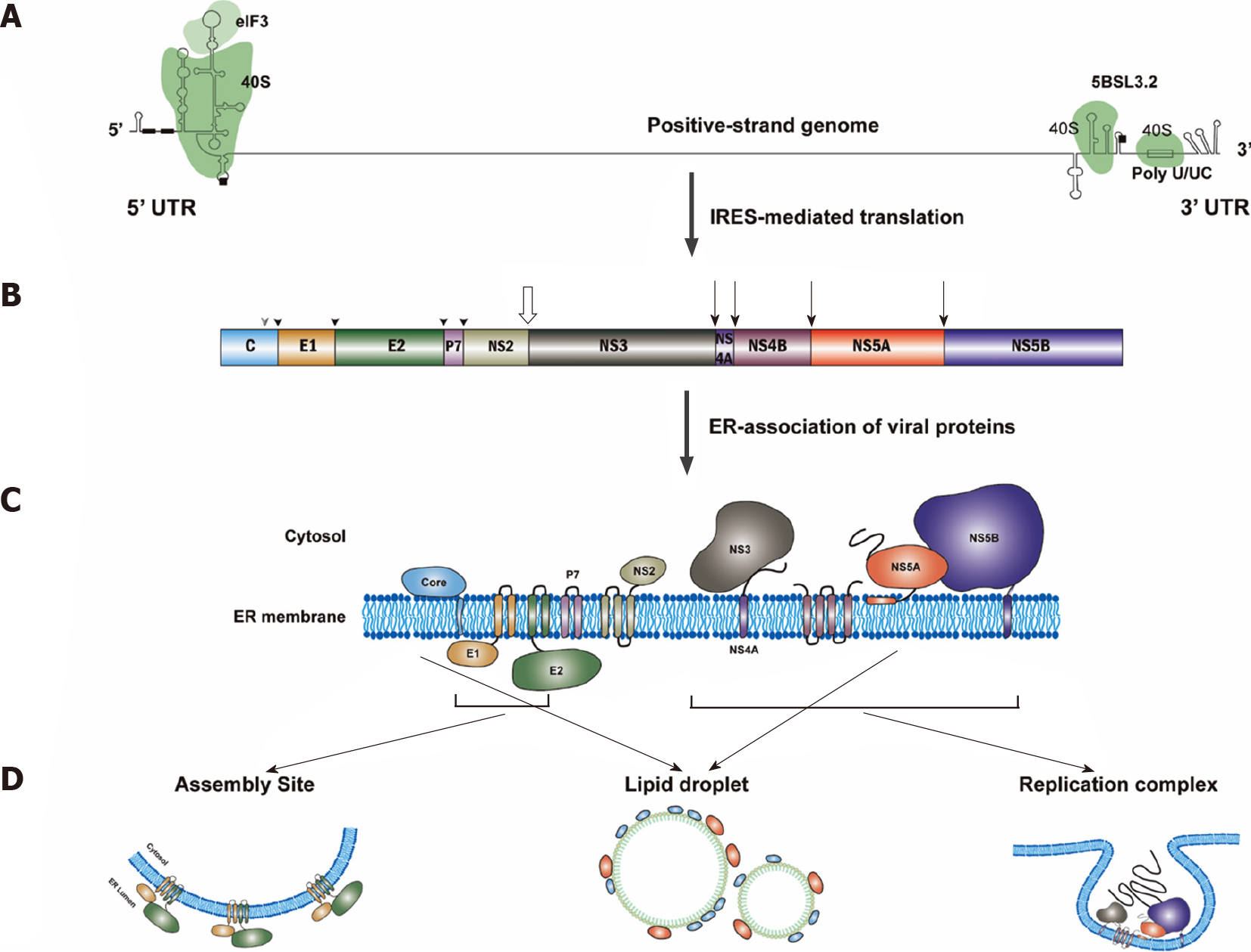
Figure 3 Hepatitis C virus protein translation.
A: Translation of hepatitis C virus (HCV) genomic RNA is regulated by the internal ribosome entry site in the 5’-untranslated region (5’UTR) along with a short segment of the core gene sequence, the cis-acting replication element in the NS5B coding region and by the entire 3’UTR. Binding sites for eIF3 and 40S were marked. The start and stop codons for protein translation are marked by black squares, while two recognition sites on the 5’UTR for miR122 are marked by black rectangles; B: Polyprotein is co- and post-translationally cleaved by host or viral proteases to yield the structural proteins (core, E1 and E2) and the nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B proteins). Core, E1 and E2 are processed by cellular signal peptidase (filled arrowhead). Mature core protein will be generated after further cleavage by signal peptide peptidase (empty arrowhead)[317]. The NS2/NS3 junction site is cleaved by the NS2–NS3 auto-protease[318] (empty arrow), and the remaining nonstructural proteins are processed by the NS3/4A proteinase[319] (filled arrow); C: All of the HCV proteins are associated with endoplasmic reticulum directly or indirectly[320]; D: Then, NS3, NS4A, NS4B, NS5A and NS5B proteins will form the replication complex. Core and NS5A proteins will be transferred to lipid droplets, while E1 and E2 proteins will stay in the assembly sites. IRES: Internal ribosome entry site; 5’UTR: 5’-untranslated region.
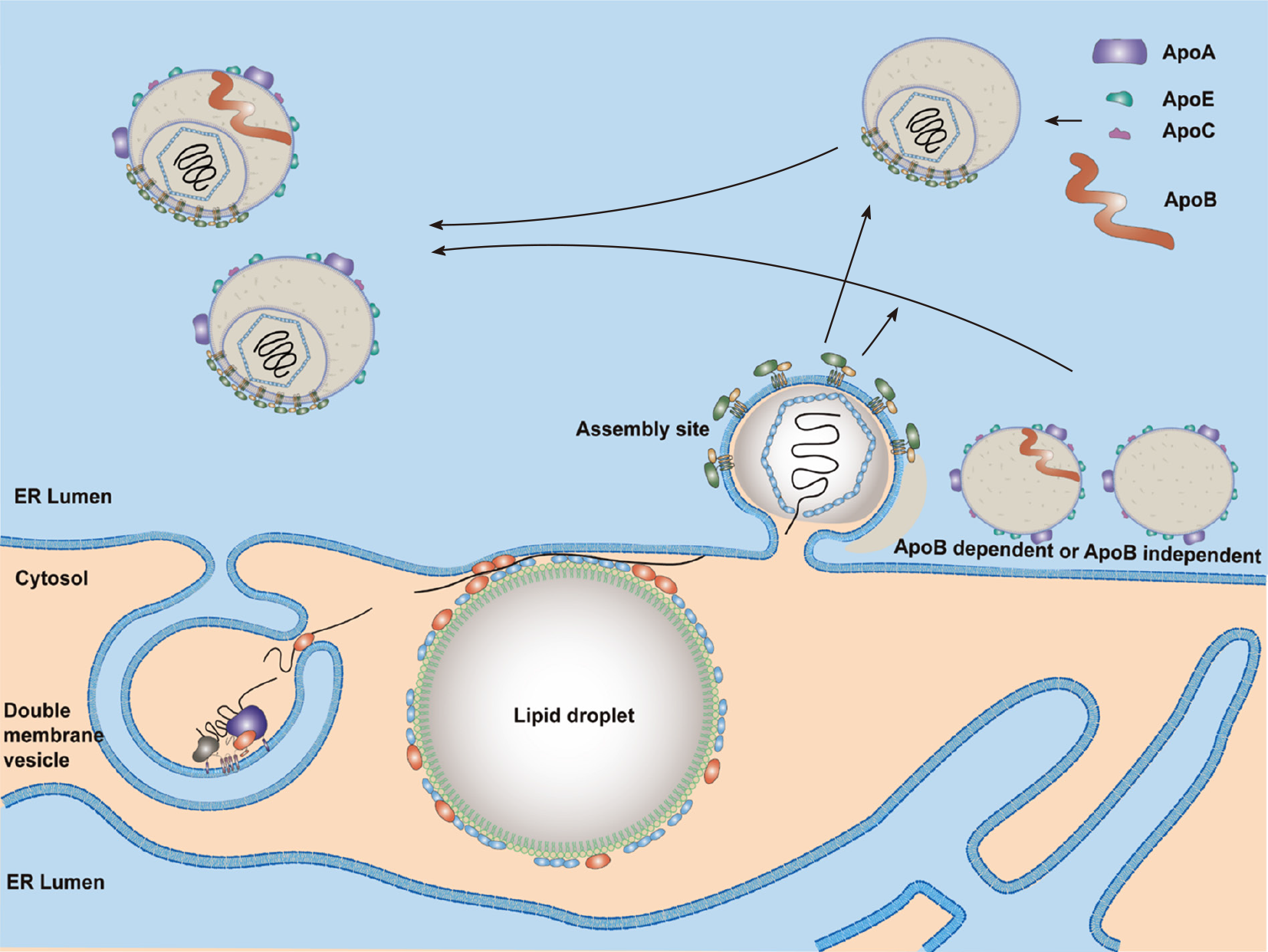
Figure 4 A proposed model for hepatitis C virus assembly.
Lipid droplets (LDs) are surrounded by endoplasmic reticulum (ER)[321]. LDs with a hepatitis C virus (HCV) core and NS5A proteins are close to replication sites [double membrane vesicle (DMV)] and assembly sites. HCV genomic RNA synthesized by the replication complex (NS3–NS5B proteins) in the DMVs will be transferred by NS5A and NS3-4A proteins and encapsidated by the core proteins to form the nucleocapsid. Then, the HCV nucleocapsid will interact with glycoproteins E1/E2 in the assembly sites and bud into the ER lumen. Both Apo-B-dependent and -independent mechanisms are possibly involved in HCV particle assembly. One model shows the production of a fused form of HCV with very-low-density lipoproteins. Another model shows the budding of HCV particles with several apolipoproteins but not Apo-B. Nascent viral particles may be further lipidated by luminal lipoproteins and incorporated with exchangeable apolipoproteins. ER: Endoplasmic reticulum.
- Citation: Li HC, Yang CH, Lo SY. Cellular factors involved in the hepatitis C virus life cycle. World J Gastroenterol 2021; 27(28): 4555-4581
- URL: https://www.wjgnet.com/1007-9327/full/v27/i28/4555.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i28.4555