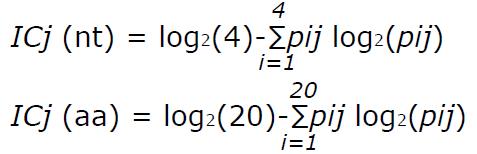Copyright
©The Author(s) 2018.
World J Gastroenterol. May 21, 2018; 24(19): 2095-2107
Published online May 21, 2018. doi: 10.3748/wjg.v24.i19.2095
Published online May 21, 2018. doi: 10.3748/wjg.v24.i19.2095
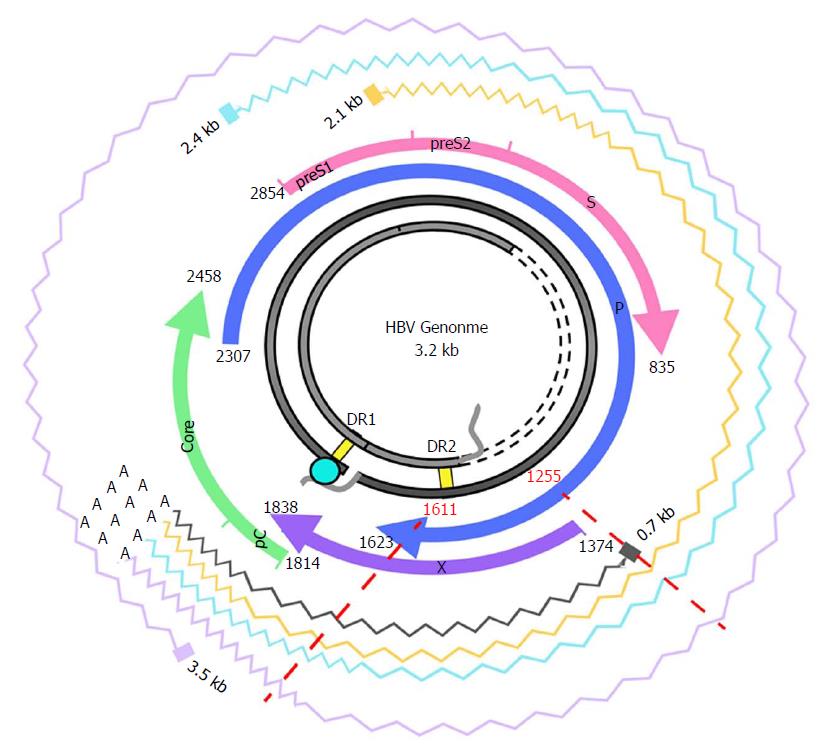
Figure 1 Hepatitis B virus genome and transcripts.
The figure shows the HBV genome (in grey), with the DR1 and DR2 (direct repeat) regions, necessary for viral DNA synthesis. The viral ORFs are highlighted with colored arrows, and nucleotide positions are reported. Wavy lines show the various HBV transcripts: the 3.5-kb transcript, corresponding to the pregenomic RNA, which is translated to the core and polymerase and later subjected to reverse-transcription in the viral capsid, or to the precore/core transcript, which is translated to the precore protein; the 2.4-kb and 2.1-kb transcripts, which are translated, respectively, to large and medium/small HBsAg); and the 0.7-kB transcript, which is translated to the HBx protein. The region analyzed in this study and its corresponding nt positions are indicated by red dashed lines. Note that the region of interest is included in all the viral transcripts. HBV: Hepatitis B virus; ORF: Open reading frame.
Math 1 Math(A1).
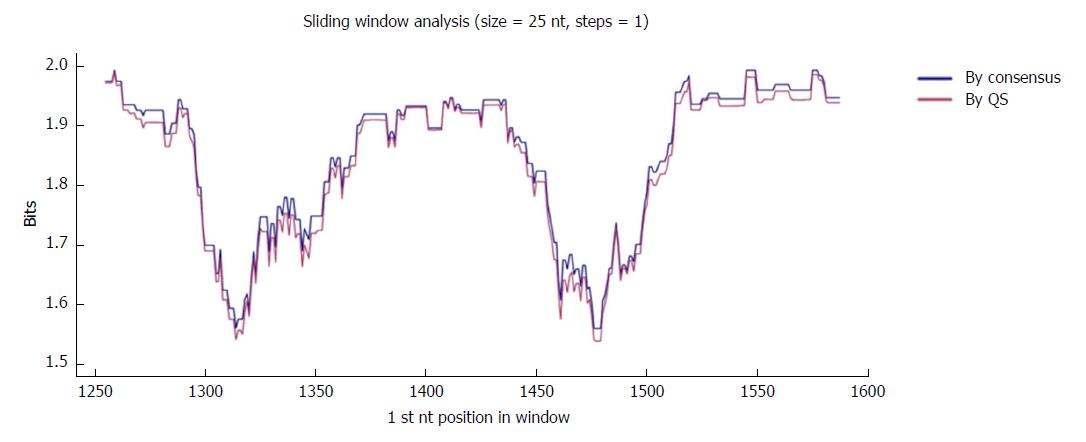
Figure 2 Sliding window analysis of the nucleotide region of interest in the hepatitis B virus X gene (nt 1255-1611).
Each point on the graph is the result of the mean information content (IC, bits) of the windows 25-nt in size, with displacement between them in 1-nt steps. The purple line represents the mean IC from the multiple alignments of all haplotypes (in abundance > 0.25%) in the quasispecies (QS) of all patients (n = 720), whereas the blue line shows the mean IC obtained from the multiple alignments of the consensus obtained for each patient (n = 27).
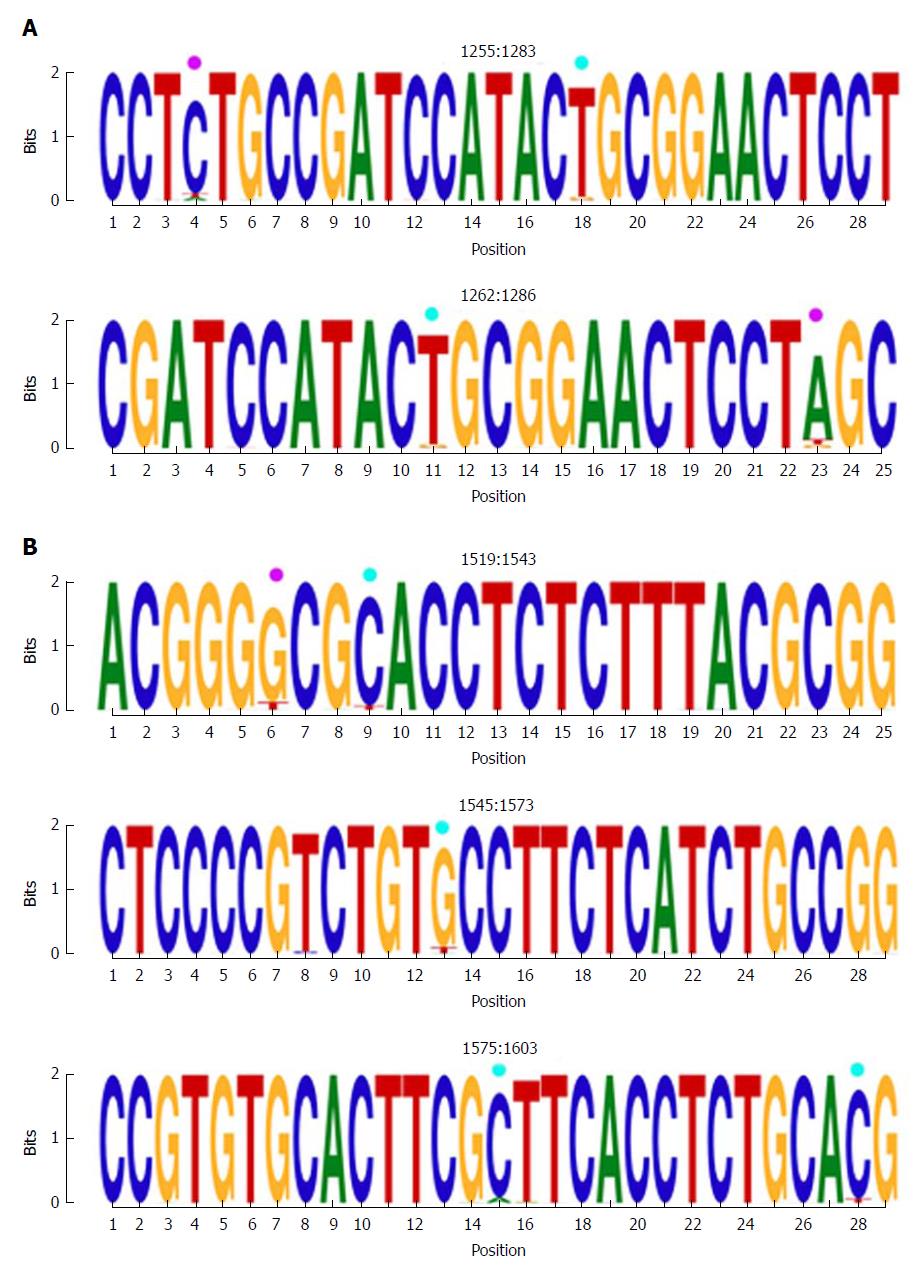
Figure 3 Representation by sequence logos of the information content of the most conserved regions detected in the multiple alignment of all nucleotide haplotypes obtained (quasispecies), nts 1255-1286 and nts 1519-1603.
The relative sizes of the letters in each stack of nt sequence logos indicate their relative frequencies at each position within the multiple alignments of nt haplotypes. The total height of each stack of letters depicts the IC of each nt position, measured in bits (Y-axis): 0 bits being the minimum and 2, the maximum conservation. Nucleotide positions with an IC between 1.6 and 1.8 (80%-90% of maximum conservation) are indicated by light blue circles and those with an IC between 1.4 and 1.6 bits (70%-80% of maximum conservation) by pink circles. IC: Information content.
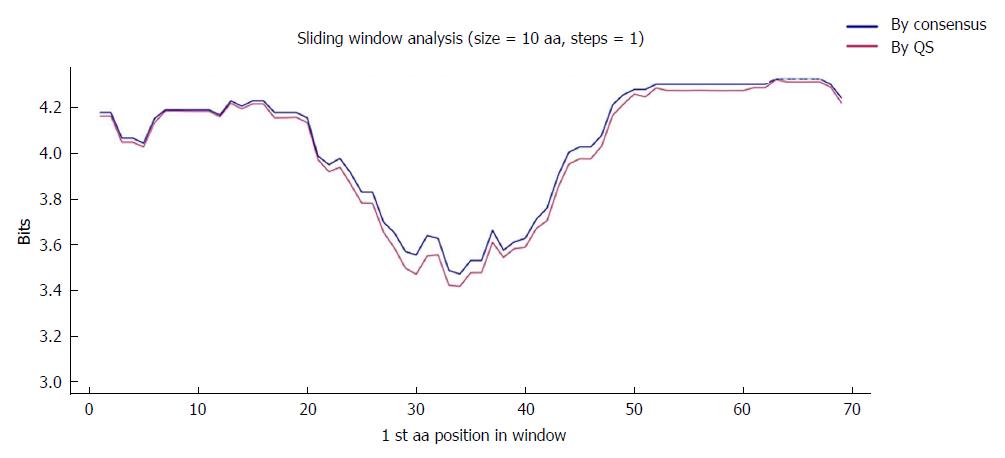
Figure 4 Sliding window of the coded the core and X protein amino acid sequence (aa 1-79).
Each point on the graph is the result of the mean information content (IC, bits) between windows of 10-aa size with displacement between them in 1-aa steps. The purple line represents the mean IC from the multiple alignments of all haplotypes (in abundance > 0.25%) in the quasispecies (QS) of all patients (n = 330), whereas the blue line shows the mean IC obtained from the multiple alignments of the consensus obtained for each patient (n = 27).
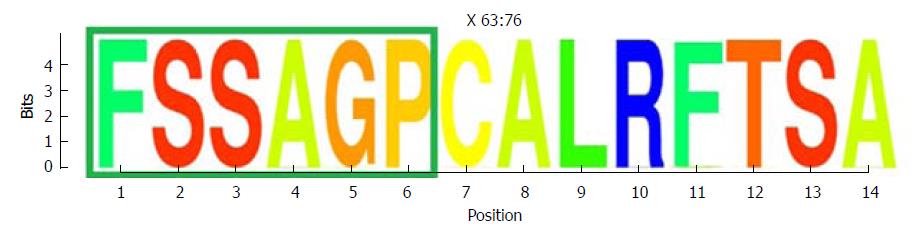
Figure 5 Representation by sequence logos of the information content of the conserved region in the the core and X protein amino acid sequence (aa 63-76).
The relative sizes of the letters in each stack in the sequence logos indicate their relative frequencies at each position within the multiple alignments of aa haplotypes obtained. The total height of each stack of letters depicts the IC of each aa position, measured in bits (Y-axis): 0 bits being the minimum and 4.32 bits the maximum conservation. Amino acids belonging to the Kunitz-like domain portion are framed in green. IC: Information content.
- Citation: González C, Tabernero D, Cortese MF, Gregori J, Casillas R, Riveiro-Barciela M, Godoy C, Sopena S, Rando A, Yll M, Lopez-Martinez R, Quer J, Esteban R, Buti M, Rodríguez-Frías F. Detection of hyper-conserved regions in hepatitis B virus X gene potentially useful for gene therapy. World J Gastroenterol 2018; 24(19): 2095-2107
- URL: https://www.wjgnet.com/1007-9327/full/v24/i19/2095.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i19.2095