Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Nov 28, 2014; 20(44): 16498-16517
Published online Nov 28, 2014. doi: 10.3748/wjg.v20.i44.16498
Published online Nov 28, 2014. doi: 10.3748/wjg.v20.i44.16498
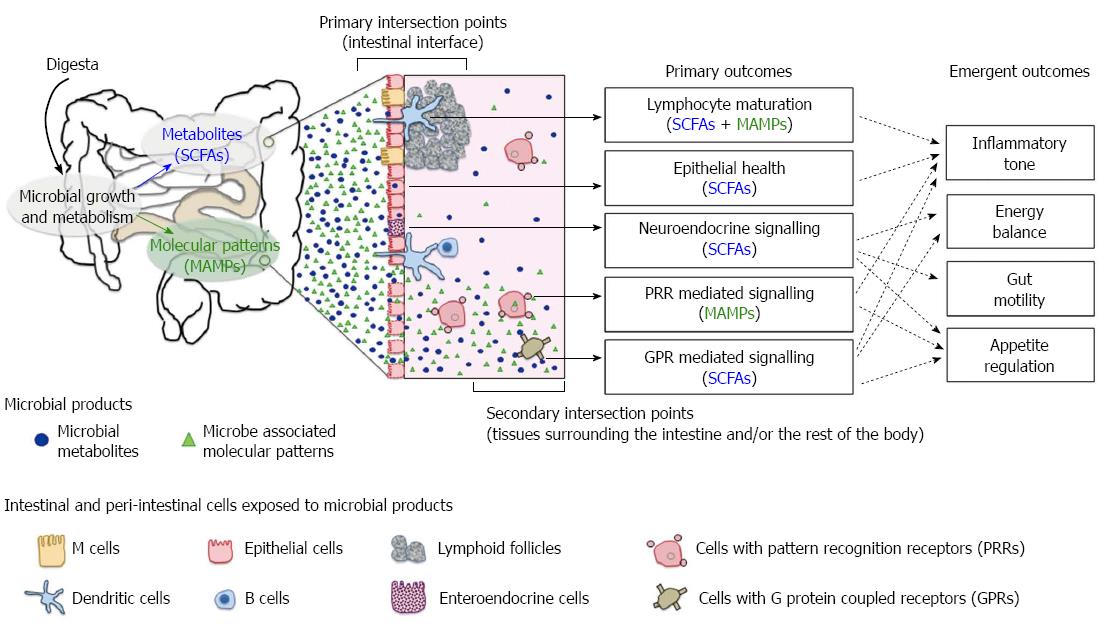
Figure 1 Axes of host-microbial interaction that influence health.
Short chain fatty acids (SCFAs) and microbe-associated molecular patterns (MAMPs) are the key microbial signals detected by the host. Outcomes of host-microbiome interactions are contingent on the microbial product involved, the type of host cells exposed to microbial signals and the location of contact. The primary intersection points occur at the intestinal epithelial interface. Sampling of luminal MAMPs and uptake of SCFAs have a direct impact on gut epithelium, lymphoid and neuroendocrine systems. The secondary intersection points occur externally to the intestinal tissues. Translocated or “escaped” microbial products can activate pattern recognition receptors (PRRs) and specific G protein coupled receptors (GPRs) on a wide range of host cells beyond the epithelium. A compromised gut barrier amplifies host-microbiome interactions in the secondary intersection points and the downstream effects of PRR and GPR signalling cascades. Host outcome is an emergent property of all axes of interactions.
Figure 2 Multiple host-mediated mechanisms regulate bacterial growth and their activities.
These pathways may act against the microbiota in a generalised manner or influence bacteria with distinct properties (blue). A: Substrates from diet are key energy sources for bacterial growth. Changes in feeding pattern will shape the microbiome structure and associated products; B: Ingestion of dietary fibre and osmotically active compounds promotes gut motility. Faster transit rate flushes out slow growing organisms and those without the ability to adhere to the intestines; C: Release of bile in response to dietary fat selects against bile-sensitive bacteria but promotes those with the capacity to obtain energy via anaerobic respiration; D: Mucin secreted by goblet cells physically prevents the penetration of bacteria into gut epithelium, and it also promotes bacteria that utilise mucin as growth substrates; E: Paneth cells in the gut epithelium secrete effector molecules with broad-spectrum antimicrobial activity, e.g. defensins, lysozyme and RegIIIγ, which contribute to the innate barrier against microbial colonisation; F: Migration of flagellated bacteria is inhibited by secretory immunoglobulin A (IgA), which facilitates the exclusion of bacteria at the epithelium; G: When mucin synthesis and release is impaired, pathobionts may penetrate the mucosal epithelium and trigger the inflammatory cascade. Byproducts of inflammation confer a growth advantage for organisms that obtain energy through anaerobic respiration.
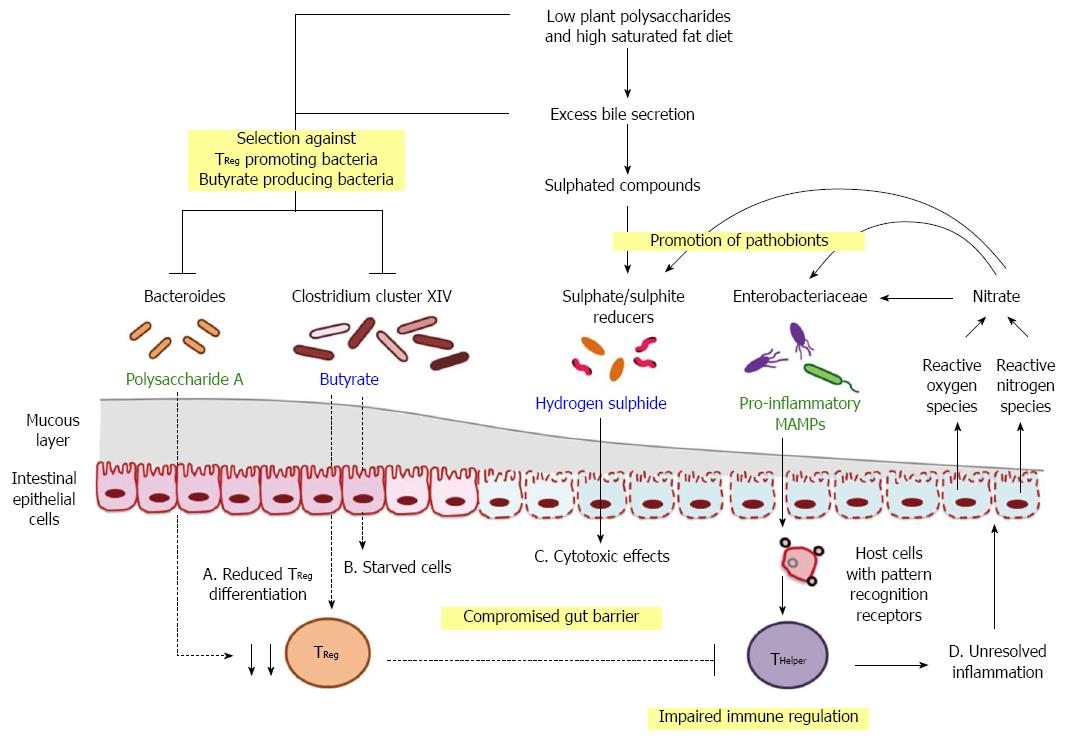
Figure 3 Hypothesised triggers and drivers in diet-induced dysbiosis.
Progression from homeostasis to clinical manifestation of metabolic dysfunction may emerge from shifts in microbe-associated molecular patterns (MAMPs; green) and metabolites (blue), initiated by long-term consumption of diets with reduced amount of dietary fibre but high saturated fat. A: Reduction in the availability of fermenting substrates in conjunction with excess secretion of anti-bacterial bile acids can alter the competition dynamics of commensal organisms and pathobionts. Consequent depletion of polysaccharide A and butyrate promotes immune dysfunction by altering the balance of regulatory T cells (TReg) and helper T cells (THelper); B and C: Shifts in microbial products contribute to the impairment of gut barrier function and the leakage of MAMPs; D: Dietary factors, microbial signals and host responses act in concert to drive inflammation, which provides a positive feedback pathway in favour of chronic disease development.
- Citation: Ha CW, Lam YY, Holmes AJ. Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health. World J Gastroenterol 2014; 20(44): 16498-16517
- URL: https://www.wjgnet.com/1007-9327/full/v20/i44/16498.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i44.16498