Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Jul 7, 2014; 20(25): 8139-8150
Published online Jul 7, 2014. doi: 10.3748/wjg.v20.i25.8139
Published online Jul 7, 2014. doi: 10.3748/wjg.v20.i25.8139
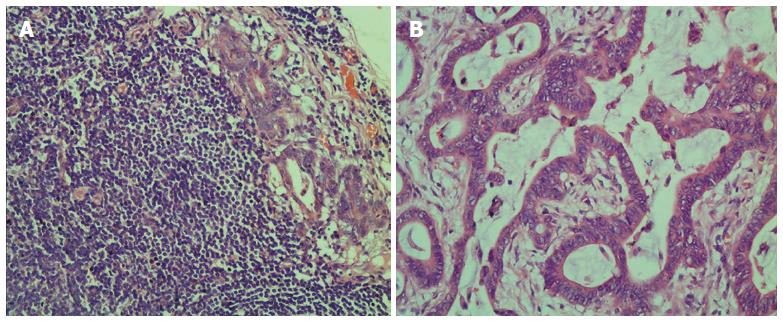
Figure 1 Metastatic lymph node and tumor tissues of colorectal cancer revealed by HE staining.
A: Metastatic lymph node; B: Tumor tissue.
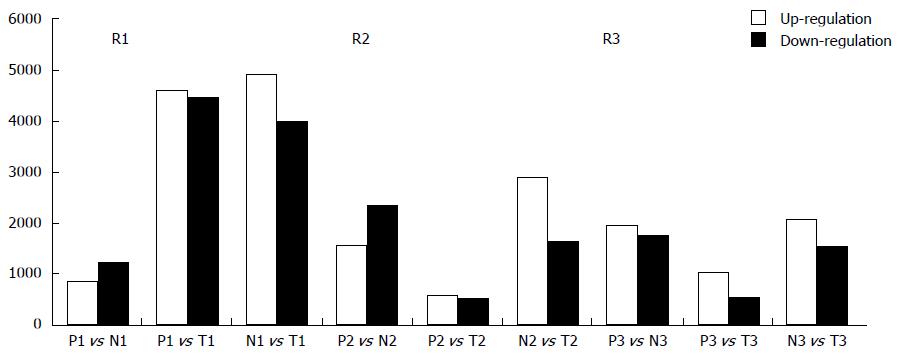
Figure 2 Counts of up-regulated and down-regulated long noncoding RNAs.
Up-regulated and down-regulated long noncoding RNAs (lncRNAs) varied in different patients when comparing different samples: many lncRNAs were determined to be significantly up-regulated or down-regulated in metastatic lymph nodes (MLN), normal lymph node (NLN) and tumor tissues of colorectal cancer (CRC) in three patients by microarray analysis. The number of differently expressed lncRNAs between NLN and tumor tissue was much larger than those of differently expressed lncRNAs both between MLN and NLN and between MLN and tumor tissue. M: Metastatic lymph nodes; N: Normal lymph nodes; T: Tumor tissue of colorectal cancer.
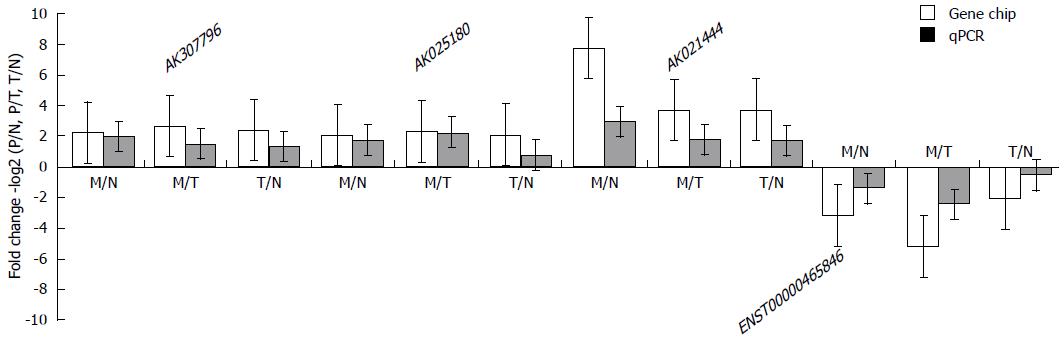
Figure 3 Comparison between microarray data and quantitative real-time polymerase chain reaction results.
AK021444, ENST00000425785, AK307796 and ENST00000465846 were determined to be differentially expressed by microarray in 3 colorectal cancer patients, and this result was validated by quantitative polymerase chain reaction result (qPCR). AK021444, ENST00000425785, and AK307796, 3 specifically up-regulated lncRNAs in metastatic lymph nodes (MLN), were confirmed to be highly expressed in MLN, while ENST00000465846 was confirmed to be down-regulated in MLN. The validation results for the four long noncoding RNAs (lncRNAs) indicated that the microarray data correlated well with the qPCR results. M: Metastatic lymph nodes; N: Normal lymph nodes; T: Tumor tissue of colorectal cancer.
- Citation: Han J, Rong LF, Shi CB, Dong XG, Wang J, Wang BL, Wen H, He ZY. Screening of lymph nodes metastasis associated lncRNAs in colorectal cancer patients. World J Gastroenterol 2014; 20(25): 8139-8150
- URL: https://www.wjgnet.com/1007-9327/full/v20/i25/8139.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i25.8139