Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Jun 28, 2014; 20(24): 7555-7569
Published online Jun 28, 2014. doi: 10.3748/wjg.v20.i24.7555
Published online Jun 28, 2014. doi: 10.3748/wjg.v20.i24.7555
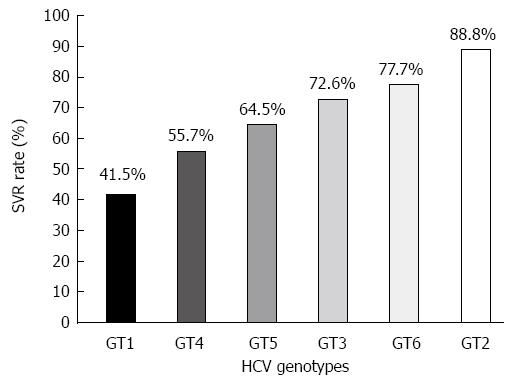
Figure 1 Average values of sustained virological response to pegylated-interferon and ribavirin combination therapy in patients infected with different hepatitis C virus genotypes.
Average of sustained virological response (SVR) rates were calculated based on SVR rates achieved in clinical studies cited in references for genotype 1 (GT1)[30,31,37,152-154], GT2[32,155-160], GT3[32,155-158,160], GT4[30,38-40,161-163], GT5[164,165] and GT6[166-172]. In those studies, standard 48-wk treatment regimens were used for GT1 and GT4, and 24-wk treatment regimens were used for GT2 and GT3. Since the treatment regimens for GT5 and GT6 have not been established yet, the treatment duration varied among different studies where it ranged from 24 to 52 wk. HCV: Hepatitis C virus.
Figure 2 Probability of different aa residues at positions 70 (A) and 91 (B) of the core protein of different hepatitis C virus subtypes.
The probability was estimated based on the core protein sequences available in hepatitis C virus (HCV) database (http://hcv.lanl.gov/content/index). The length of an aa symbol-letter represents its probability for a given HCV subtype. R: Arginine; Q: Glutamine; H: Histidine; L: Leucine; C: Cysteine; M: Methionine.
Figure 3 Sequence alignment of interferon sensitivity-determining region of major hepatitis C virus subtypes.
References of aligned sequences are: hepatitis C virus (HCV)-1b, Enomoto et al[90]; HCV-1a, AF009606; HCV-2a and -2b, Murakami et al[104]; HCV-3a, GU814263; HCV-4a, El-Shamy et al[73]; HCV-5a, AF064490; HCV-6a, DQ480512. ISDR: Interferon sensitivity-determining region.
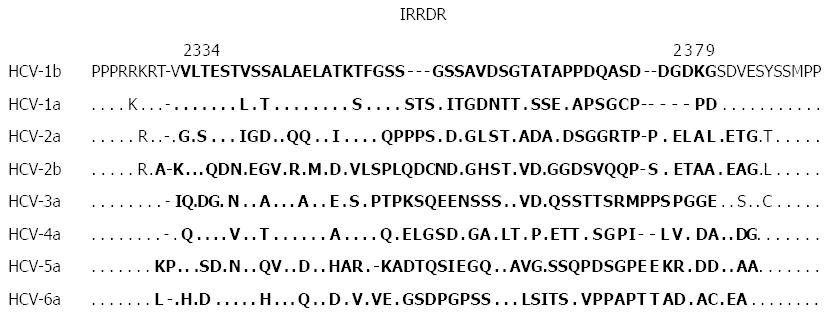
Figure 4 Sequence alignment of interferon/ribavirin resistance-determining region and its vicinity of major hepatitis C virus subtypes.
The residues in interferon/ribavirin resistance-determining region (IRRDR) are written in bold face letters. References of aligned sequences are: hepatitis C virus (HCV)-1b, El-Shamy et al[122]; HCV-1a, AF009606; HCV-2a and -2b, Murakami et al[104]; HCV-3a, GU814263; HCV-4a, El-Shamy et al[73]; HCV-5a, AF064490; HCV-6a, DQ480512.
- Citation: El-Shamy A, Hotta H. Impact of hepatitis C virus heterogeneity on interferon sensitivity: An overview. World J Gastroenterol 2014; 20(24): 7555-7569
- URL: https://www.wjgnet.com/1007-9327/full/v20/i24/7555.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i24.7555