Copyright
©2014 Baishideng Publishing Group Co.
World J Gastroenterol. Apr 28, 2014; 20(16): 4761-4770
Published online Apr 28, 2014. doi: 10.3748/wjg.v20.i16.4761
Published online Apr 28, 2014. doi: 10.3748/wjg.v20.i16.4761
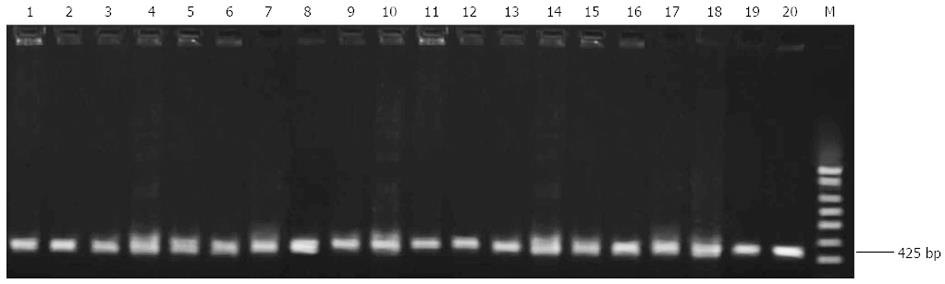
Figure 1 PCR amplification of the 23S rRNA gene of Helicobacter pylori strains.
1-10: Randomly selected clarithromycin-sensitive strains; 11-20: Randomly selected clarithromycin-resistant strains; M: 100 bp DNA ladder.
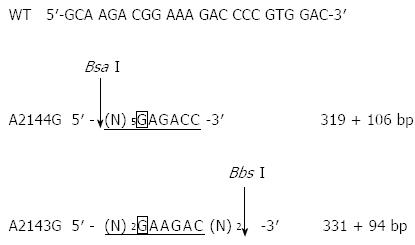
Figure 2 Two new restriction sites for Bbs I and Bsa I caused by clarithromycin resistance-associated mutations A2143G and A2144G in the 23S rRNA gene of Helicobacter pylori strains.
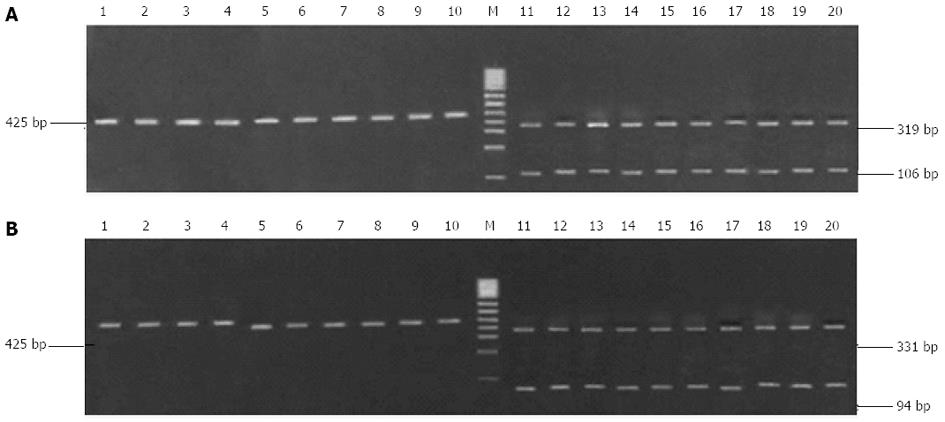
Figure 3 Digestion of the amplified fragment of the 23S rRNA gene from Helicobacter pylori isolates with restriction endonuclease Bbs I (A) and Bsa I (B).
1-10: Randomly selected clarithromycin-sensitive strains; 11-20: Randomly selected clarithromycin-resistant strains; M: 100 bp DNA ladder.
Figure 4 Comparison of the 23S rRNA gene sequence of Helicobacter pylori isolates with that of HPJ99 strain.
S: A clarithromycin-sensitive isolate; R1-5: clarithromycin-resistant strains.
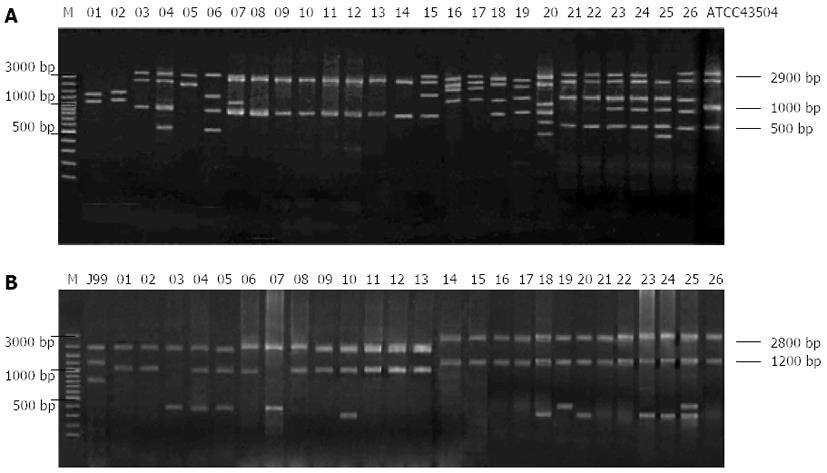
Figure 5 REP-PCR analysis of 26 clarithromycin-resistant (A) and clarithromycin-sensitive (B) Helicobacter pylori isolates.
M: λDNA/EcoR I-Hind III marker; 1-26: Clarithromycin-resistant Helicobacter pylori isolates; ATCC43504: A standard Helicobacter pylori strain.
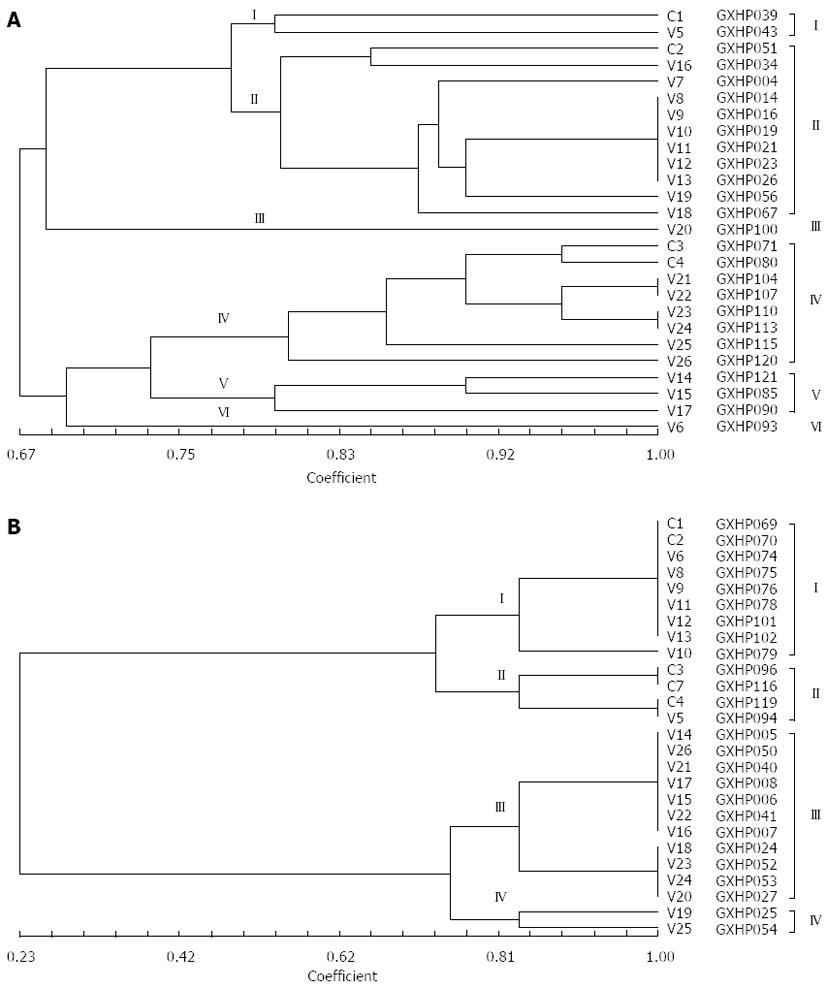
Figure 6 Dendrogram of REP-PCR DNA fingerprints for clarithromycin-resistant Helicobacter pylori strains.
A: 6 genotypes; B: 4 genotypes.
-
Citation: Zhao LJ, Huang YQ, Chen BP, Mo XQ, Huang ZS, Huang XF, Wei LD, Wei HY, Chen YH, Tang HY, Huang GR, Qin YC, Li XH, Wang LY.
Helicobacter pylori isolates from ethnic minority patients in Guangxi: Resistance rates, mechanisms, and genotype. World J Gastroenterol 2014; 20(16): 4761-4770 - URL: https://www.wjgnet.com/1007-9327/full/v20/i16/4761.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i16.4761