Copyright
©2012 Baishideng Publishing Group Co.
World J Gastroenterol. Feb 14, 2012; 18(6): 576-582
Published online Feb 14, 2012. doi: 10.3748/wjg.v18.i6.576
Published online Feb 14, 2012. doi: 10.3748/wjg.v18.i6.576
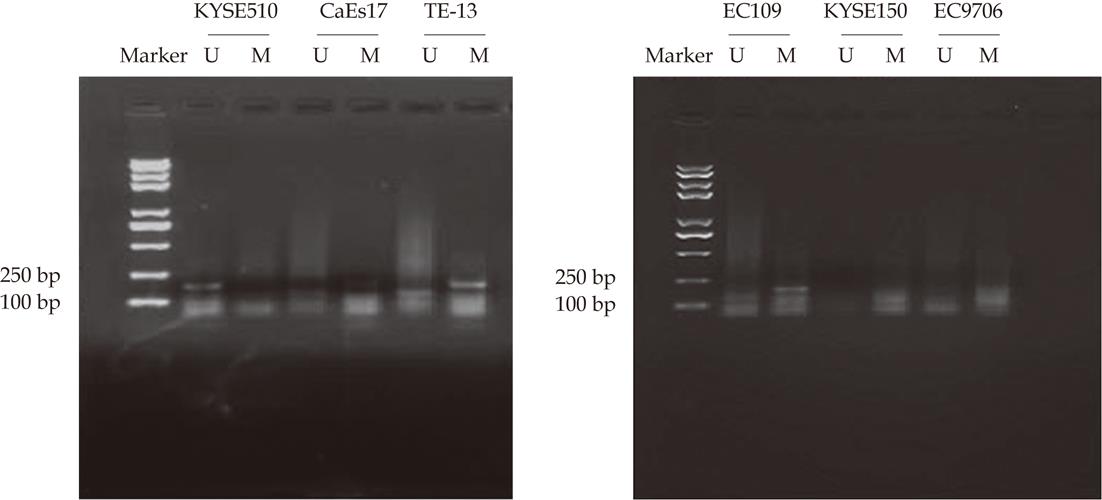
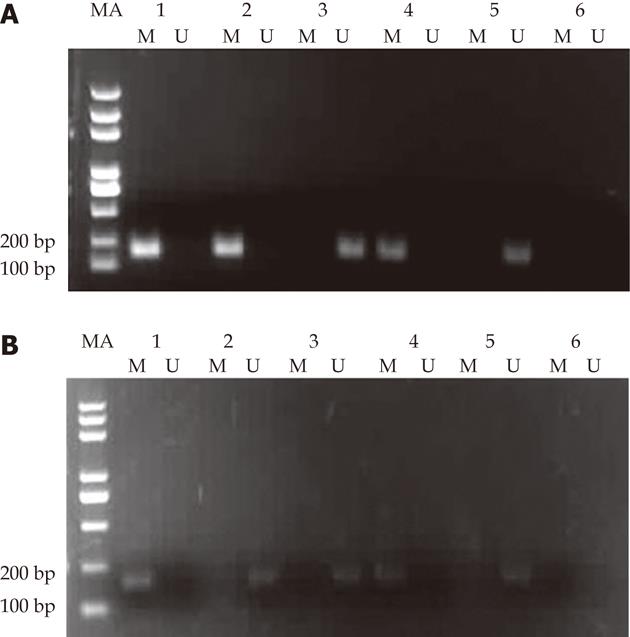
Figure 1 Methylation-specific polymerase chain reaction analyses of retinoblastoma protein-interacting zinc finger 1 promoter methylation status using genomic DNA extracted from 6 human esophageal squamous cell carcinoma cell lines, products of 177 bp and 175 bp were expected for methylated (M) and unmethylated (U) DNA.
M: Methylation-specific amplification (177 bp); U: Unmethylation-specific amplification (175 bp).
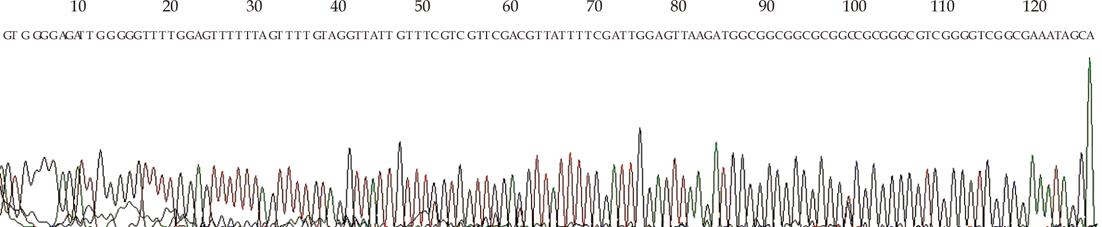
Figure 2 Sequencing analysis of methylated (M) polymerase chain reaction products of methylation-specific polymerase chain reaction from TE13
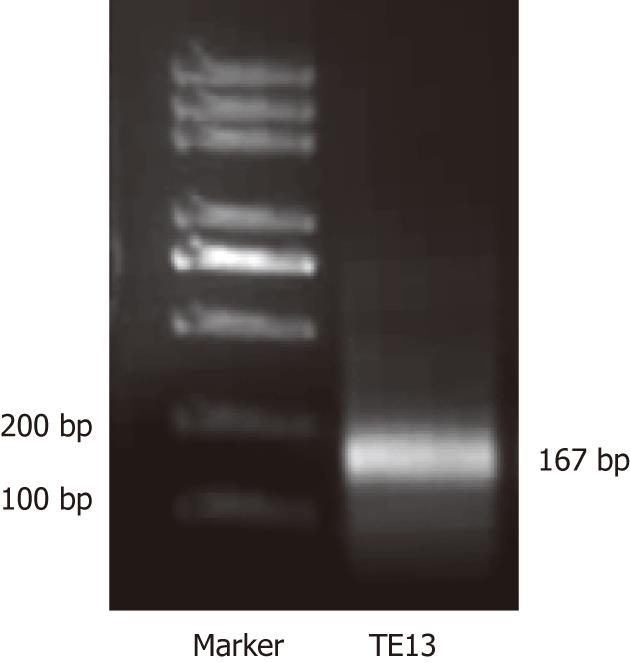
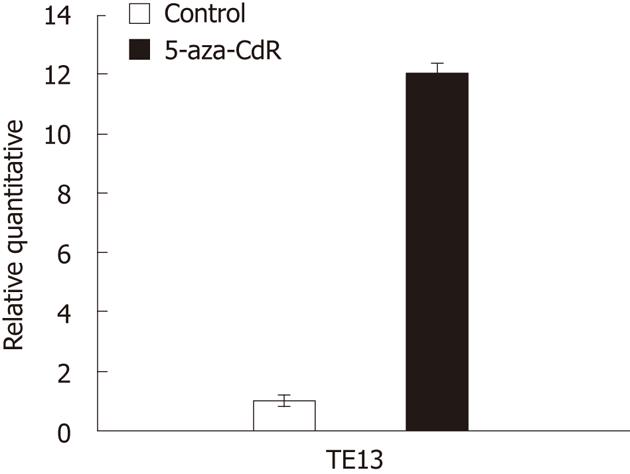
Figure 3 Reverse transcription-polymerase chain reaction for RIZ1 expression in TE13 treated or not by 5-aza-CdR.
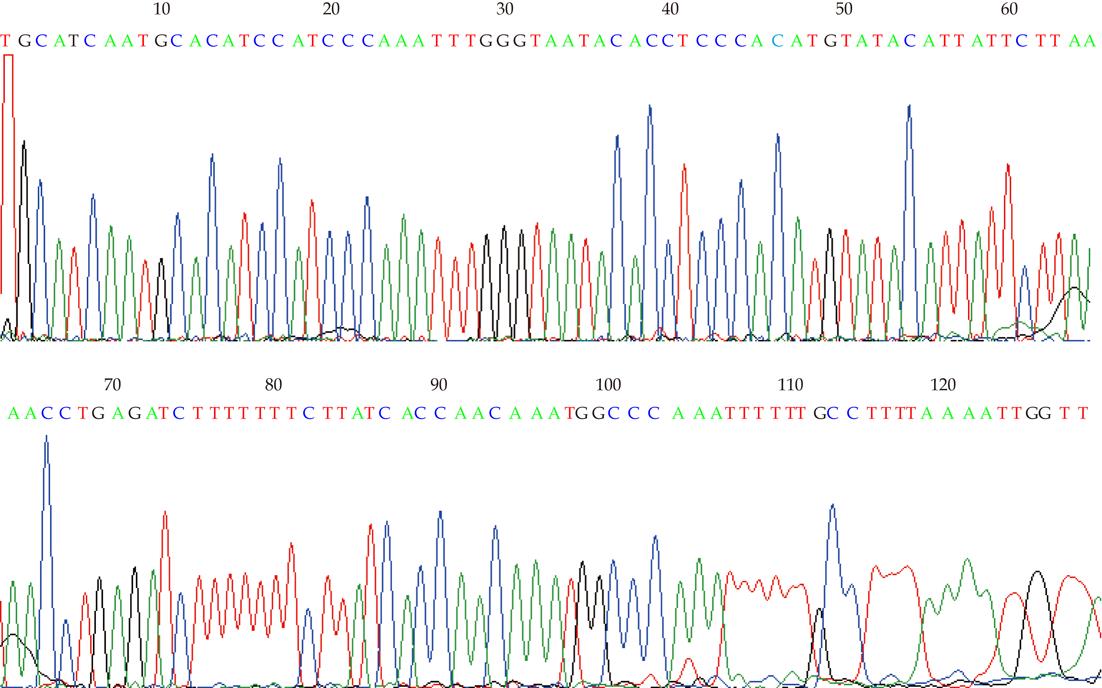
Figure 4 Sequencing analysis of reverse transcription-polymerase chain reaction production using retinoblastoma protein-interacting zinc finger gene 1 F-primer, which is the fragment aimed at.
Figure 5 The expression of retinoblastoma protein-interacting zinc finger 1 when the drug 5-aza-CdR is used.
Figure 6 Methylation-specific polymerase chain reaction electrophoresis for cancerous, neighboring and distal ending normal tissues.
A: The results of electrophoresis for the polymerase chain reaction (MSP) products of positive retinoblastoma protein-interacting zinc finger gene 1 (RIZ1) methylation amplification sufferers in both their cancerous and neighboring normal tissues; B: The results of electrophoresis for the MSP products of sufferers in whose cancerous tissue positive RIZ1 methylation amplification appears while positive unmethylation amplification is present in the corresponding neighboring normal tissues. MA: Marker; M: Methylation (177bp); U: Unmethylation (175bp);1: Carcinomas; 2: Matched neighboring normal esophageal tissues; 3: Distal ending normal esophageal tissues; 4: Postive control for methylation; 5: Postive control for unmethylation; 6: Negative control.
-
Citation: Dong SW, Zhang P, Liu YM, Cui YT, Wang S, Liang SJ, He Z, Sun P, Wang YG. Study on
RIZ1 gene promoter methylation status in human esophageal squamous cell carcinoma. World J Gastroenterol 2012; 18(6): 576-582 - URL: https://www.wjgnet.com/1007-9327/full/v18/i6/576.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i6.576