Copyright
©The Author(s) 2004.
World J Gastroenterol. Jul 15, 2004; 10(14): 2078-2081
Published online Jul 15, 2004. doi: 10.3748/wjg.v10.i14.2078
Published online Jul 15, 2004. doi: 10.3748/wjg.v10.i14.2078
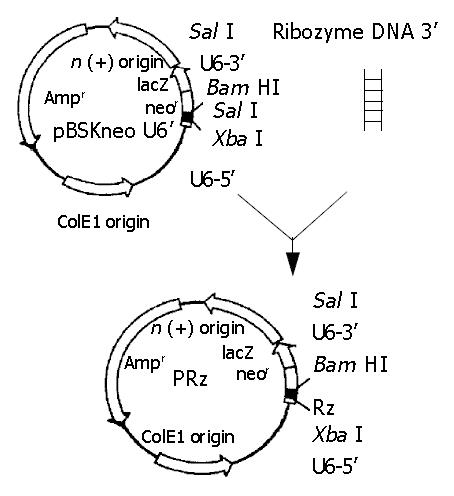
Figure 1 Ribozyme gene cloning.
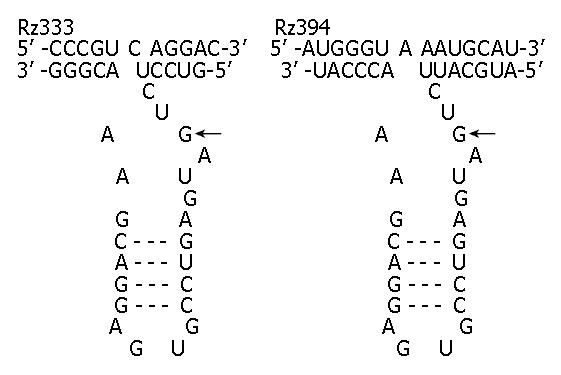
Figure 2 Sequences, structures and targets of ribozymes against caspase-7.
Hammerhead ribozymes (bottom strand) binding to their targets (top strand) to form a typical three-stem struc-ture that leads to the cleavage of caspase-7 RNA at the targets of GUC (Rz333) and GUA (Rz394).
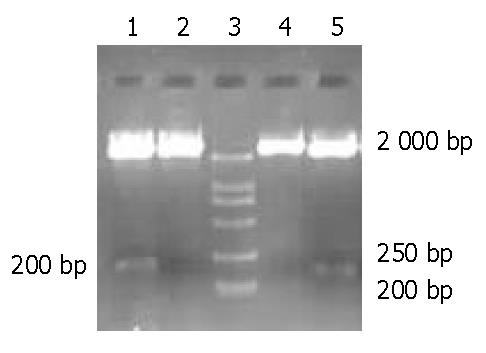
Figure 3 Restriction enzyme cleavage of reconstructed ribozyme plasmid.
Lanes 1, 2: pU6Rz333 digested by Sal I; lanes 4, 5: pU6Rz394 digested by Sal I; lane 3: Marker (DL2000).
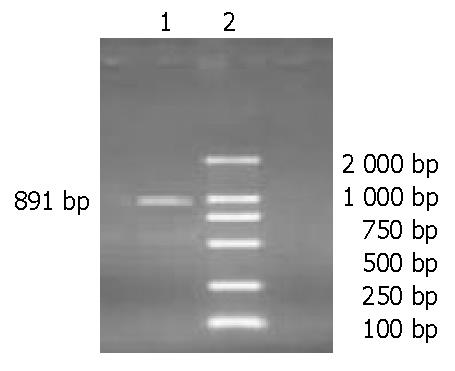
Figure 4 Agarose gel electrophoresis of RT-PCR products.
Lane 1: A 891-bp caspase-7 gene segment; lane 2: Marker (DL2000).
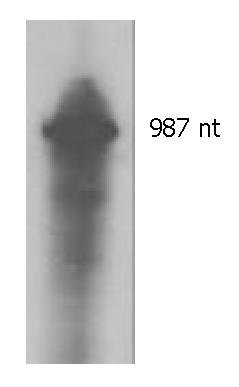
Figure 5 In vitro transcription of target RNA.
The transcript is 987 nt.
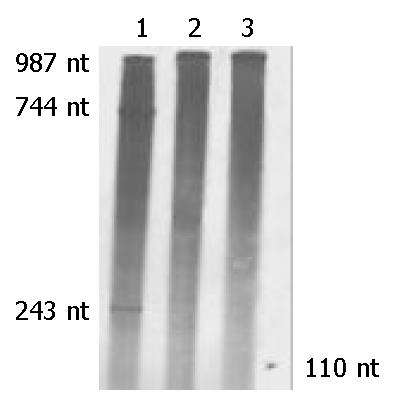
Figure 6 In vitro cleavage experiments of anti-caspase-7 ribozymes.
Lane 1: Caspase-7 mRNA was mixed with Rz333 and two segments of 243 nt and 744 nt produced by cleavage reaction. Lane 2: Caspase-7 mRNA was mixed with Rz394 and no segment was seen after cleavage. Lane 3: Caspase-7 mRNA was mixed with neither Rz333 nor Rz394. The 110-nt marker is indicated by a short arrow.
-
Citation: Zhang W, Xie Q, Zhou XQ, Jiang S, Jin YX. Expression and
in vitro cleavage activity of anti-caspase-7 hammerhead ribozymes. World J Gastroenterol 2004; 10(14): 2078-2081 - URL: https://www.wjgnet.com/1007-9327/full/v10/i14/2078.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i14.2078