Copyright
©The Author(s) 2004.
World J Gastroenterol. May 15, 2004; 10(10): 1447-1451
Published online May 15, 2004. doi: 10.3748/wjg.v10.i10.1447
Published online May 15, 2004. doi: 10.3748/wjg.v10.i10.1447
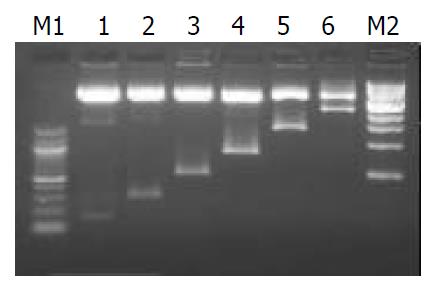
Figure 1 Electrophoresis of six constructs digested with Mlu I and Xho I.
M1: 100 bp DNA ladder marker, M2: 1 kb DNA ladder marker, Lane1: pCOLH10.1, lane2: pCOLH10.27, lane3: pCOLH10.5, Lane4: pCOLH10.9, Lane5: pCOLH11.5, Lane6: pCOLH12.5. Six recombinant plasmids containing serial 5’-de-leted flanking sequences of human α1(I) procollagen gene as putative promoter were digested with Mlu I and Xho I at 37 °C for 1 h. The digested DNAs were fractionated on 15 g/L agar-ose gel showing vector DNA (4.3 kb) and insertion promoters with different sizes.
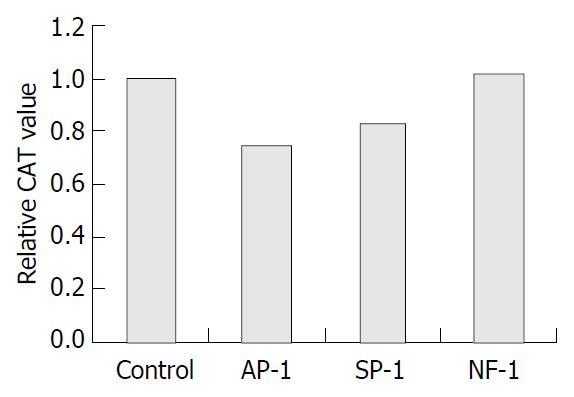
Figure 2 Effects of consensus DNA on CAT expression in pCOLH10.
27 transfected cells. Control: mock DNA transfection, Ap-1: transfection of Ap-1 consensus DNA (10 μg), Sp-1: trans-fection of Sp-1 consensus DNA (10 μg), NF-1: transfection of NF-1 consensus DNA (10 μg). For the cells transfected with pCOLH10.27, transfection of consensus recognition DNA for Ap-1, Sp-1 or NF-1 was performed 24 h after initial transfection. Reporter gene CAT was determined with ELISA after another 24 h. Relative CAT values in different transfection groups were calculated relative to that in mock DNA transfection. The result represented three independent experiments. aP < 0.05 compared to control.
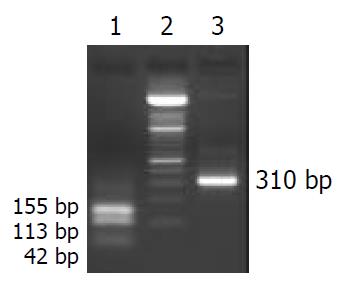
Figure 3 Electrophoresis (20 g/L agarose gel) of the fragment -268 to +42 bp digested with EcoR II.
Lane1: EcoR II-digested fragment, Lane2: 100 bp DNA ladder, Lane3: 310 bp length of fragment spanning from -268 to +42 bp. The 310 bp fragment spanning -268 to +42 bp of the human α1(I) procollagen gene was obtained by PCR with p5.3Kα1 as template. The fragment was digested with EcoR II. Electrophoresis (20 g/L agarose gel) of the digested mixture showed 3 bands with different sizes (42 bp, 113 bp, 155 bp) which were labeled and used as probes in EMSA.
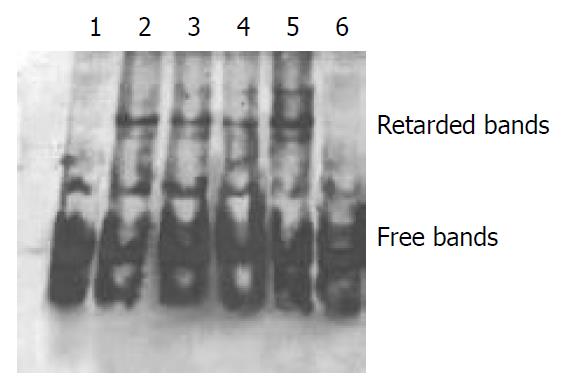
Figure 4 Result of EMSA with EcoR II-digested -268 to +42 bp as probe.
Lane1: Labeled DNA, Lane2: Labeled DNA+ nuclear protein +NF-1 consensus DNA, Lane3: Labeled DNA + nuclear protein + Sp-1 consensus DNA, Lane4: Labeled DNA + nuclear protein + Ap-1 consensus DNA, Lane 5: Labeled DNA + nuclear protein, Lane 6: Labeled DNA + excess unlabeled DNA + nuclear protein. The existence of several retarded bands in EMSA indi-cated that there were several nuclear protein binding sites in sequence –268 to +42 bp (lane 5). No retardation occurred when excess unlabeled DNA probe was added to the DNA-protein reaction, confirming the specificity of the retardation (lane 6). Consensus DNAs for Sp1, Ap-1 and NF-1 were among the pos-sible regulatory elements since the molar excess of the consen-sus unlabeled probe (Sp1, Ap-1, NF-1) inhibited partially the formation of retardation bands differently (lanes 2, 3 and 4).
- Citation: Gao CF, Wang H, Wang AH, Wan WD, Wu YA, Kong XT. Transcriptional regulation of human α1(I) procollagen gene in dermal fibroblasts. World J Gastroenterol 2004; 10(10): 1447-1451
- URL: https://www.wjgnet.com/1007-9327/full/v10/i10/1447.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i10.1447