Copyright
©The Author(s) 2015.
World J Transl Med. Apr 12, 2015; 4(1): 11-24
Published online Apr 12, 2015. doi: 10.5528/wjtm.v4.i1.11
Published online Apr 12, 2015. doi: 10.5528/wjtm.v4.i1.11
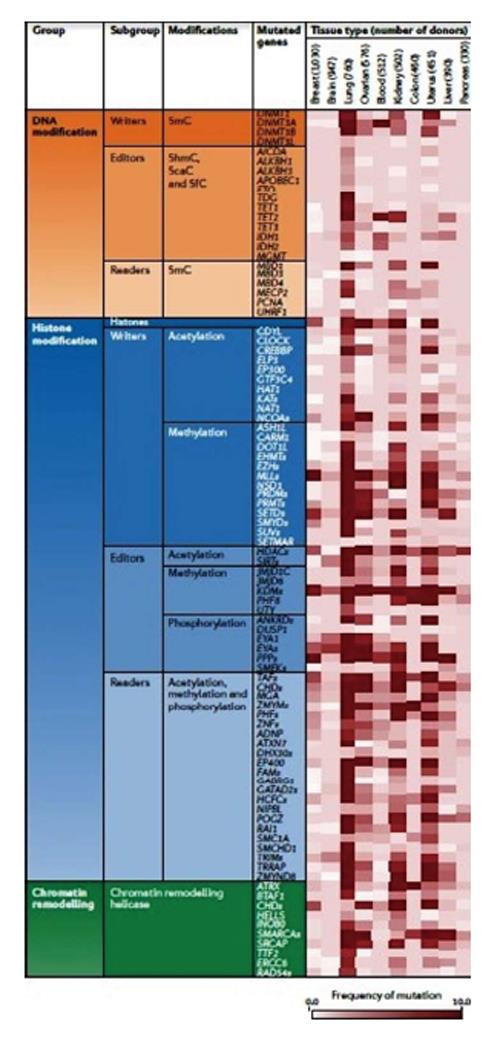
Figure 10 Some of the most known mutated genes classified in groups based on DNA modification, histone modification and chromatin remodelling enzymes.
The number of analysed tumour tissues is given. Several epigenetic enzymes present high frequencies of mutations in distinct tumor types. These data are not adjusted for chromosomal instability or mutator phenotypes, hence the frequencies reflect a combination of probable driver mutations in epigenetic regulators and the background mutation rate for the tumour type. 5caC: 5 carboxylcytosine; 5fC: 5 formylcytosine; 5hmC: 5 hydroxymethylcytosine; 5mC: 5 methylcytosine. Adapted by Plass et al[42], 2012.
- Citation: Lattanzio L, Lo Nigro C. Epigenetics and DNA methylation in cancer. World J Transl Med 2015; 4(1): 11-24
- URL: https://www.wjgnet.com/2220-6132/full/v4/i1/11.htm
- DOI: https://dx.doi.org/10.5528/wjtm.v4.i1.11