Copyright
©The Author(s) 2015.
World J Transl Med. Apr 12, 2015; 4(1): 11-24
Published online Apr 12, 2015. doi: 10.5528/wjtm.v4.i1.11
Published online Apr 12, 2015. doi: 10.5528/wjtm.v4.i1.11
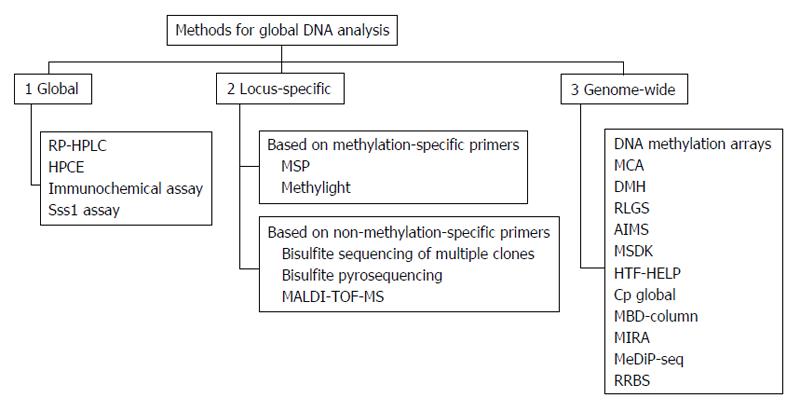
Figure 9 Methods for genomic DNA methylation analysis, classified as global, locus-specific and genome-wide.
In the case of locus-specific approaches, techniques were divided depending on the use of methylation-specific primers or not. RP-HPLC: Reverse-Phase high-performance liquid chromatography; HPCE: High performance capillary electrophoresis; Sss1 assay: Methyl group acceptance assay; MSP: Methylation-specific polymerase chain reaction (PCR); MALDI-TOF-MS: Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; MCA: Methylation CpG island amplification; DMH: Differential methylation hybridization; RLGS: Restriction-landmark genomic scanning; AIMS: Amplification of inter-methylated sites; MSDK: Methylation-specific digital karyotyping; HTF-HELP: HPAII tiny fragment enrichment by ligation-mediated PCR; MBD-column: Methylated DNA binding column; MIRA: Methylated CpG island recovery assay; MeDIP-seq: Methyl-DNA immunoprecipitation and sequencing; RRBS: Reduced Representation Bisulfite Sequencing. Adapted by Toraño et al[33], 2011.
- Citation: Lattanzio L, Lo Nigro C. Epigenetics and DNA methylation in cancer. World J Transl Med 2015; 4(1): 11-24
- URL: https://www.wjgnet.com/2220-6132/full/v4/i1/11.htm
- DOI: https://dx.doi.org/10.5528/wjtm.v4.i1.11