Copyright
©2014 Baishideng Publishing Group Inc.
World J Med Genet. May 27, 2014; 4(2): 6-18
Published online May 27, 2014. doi: 10.5496/wjmg.v4.i2.6
Published online May 27, 2014. doi: 10.5496/wjmg.v4.i2.6
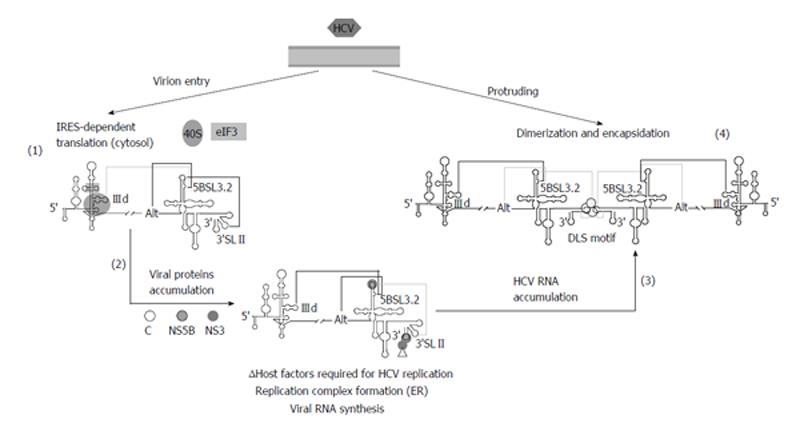
Figure 5 Proposed model for the participation of long-distant RNA-RNA interactions in the consecution of the hepatitis C virus infective cycle.
Virions penetrate inside the cell and the genomic RNA is released in the cytoplasm to initiate viral translation (1). The 40S ribosomal subunit binds to the IIId subdomain of the internal ribosome entry site (IRES) region and avoids its interaction with domain 5BSL3.2, thus favoring the contacts Alt-5BSL3.2-3’SLII. Once protein levels have surpassed a certain concentration (2), replication complexes are fixed on the surface of the endoplasmic reticulum (ER) to initiate the genomic RNA amplification. The recruitment of protein factors at the 3’X-tail would hide the 3’SLII domain. This would displace the interactions balance toward the IIId-5BSL3.2 contact, thus impeding efficient translation. Importantly, a considerable pool of molecules should alternatively display a preferred folding defined by the interaction Alt-5BSL3.2, which is indispensable for replication. The accumulation of newly naked viral genomes (3) could induce a replication-defective state by the acquisition of a favored conformation involving the interaction IIId-5BSL3.2, which would yield the dimer linkage sequence (DLS) exposed in an apical loop. The presence of the viral core chaperone protein would participate in the formation of dimeric genomes (4), which would be finally encapsidated, enveloped and released to the extracellular medium. Improbable contacts are indicated by grey lines at each step. Figure adapted from[31].
- Citation: Romero-López C, Berzal-Herranz A. Structure-function relationship in viral RNA genomes: The case of hepatitis C virus. World J Med Genet 2014; 4(2): 6-18
- URL: https://www.wjgnet.com/2220-3184/full/v4/i2/6.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v4.i2.6