Copyright
©The Author(s) 2016.
World J Biol Chem. Feb 26, 2016; 7(1): 188-205
Published online Feb 26, 2016. doi: 10.4331/wjbc.v7.i1.188
Published online Feb 26, 2016. doi: 10.4331/wjbc.v7.i1.188
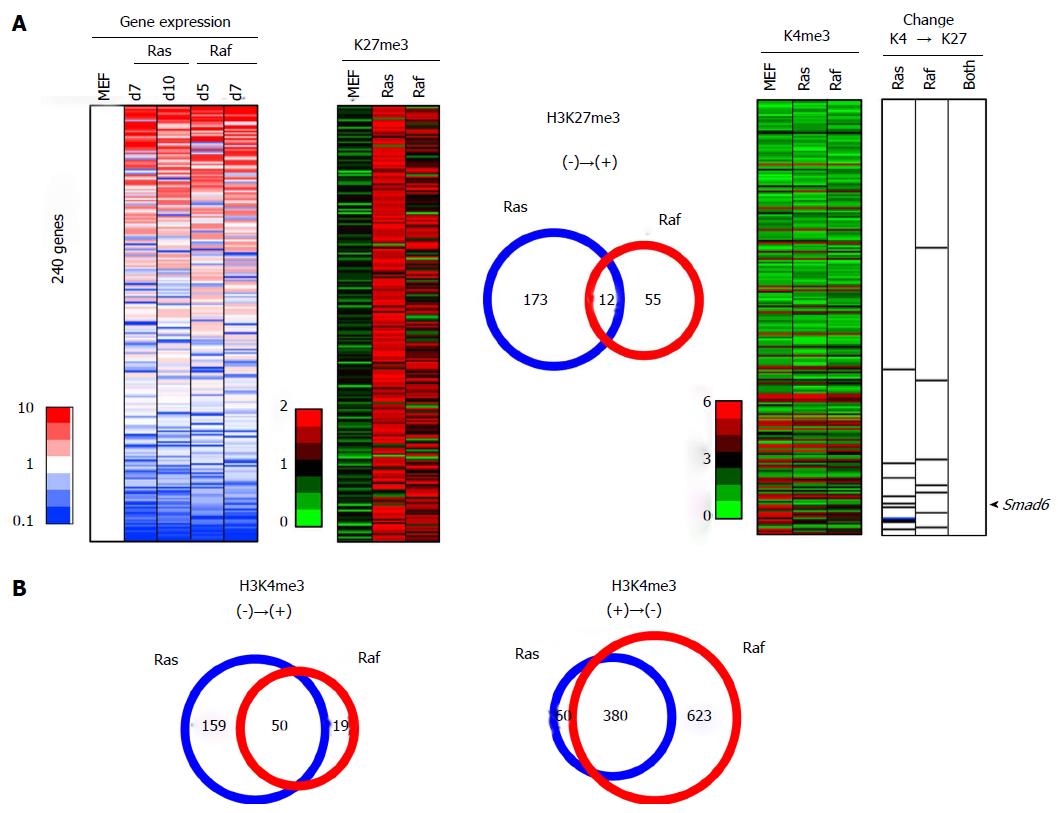
Figure 10 Integrated analysis of epigenetic and expression changes for genes acquiring H3K27me3.
A: A total of 240 genes showed < 1.0 reads per 1000000 reads in MEF, but increased to > 1.5 in both or either of Ras- and Raf-induced senescence, and were sorted by the fold expression change between MEF and mean of RasG12V cells and RafV600E cells (left). Among these 240 genes acquiring de novo H3K27me3 in senescence, 10 genes in Ras-induced senescence and 7 genes in Raf-induced senescence showed a simultaneous gain of H3K27me3 and loss of H3K4me3 (right). These genes showed a significant enrichment in downregulated genes among the 240 genes (P = 4 × 10-6 in Ras-induced senescence, and P = 0.02 in Raf-induced senescence, Kolmogorov-Smirnov test). However, these 10 genes, e.g., Smad6 (arrow head), and 7 genes did not overlap at all. Regarding the overlap of H3K27me3 alterations (center), 185 genes with H3K27me3 gain in Ras-induced senescence and 67 genes with H3K27me3 gain in Raf-induced senescence showed limited overlap (12 genes only, phi coefficient = 0.103), and this overlap in H3K27me3 gain was markedly rare compared with the overlap in H3K27me3 loss (Figure 9) (P < 1 × 10-15, log linear analysis); B: Overlap of H3K4me3 alterations. Genes with H3K4me3 gain in Ras- and Raf induced senescence overlapped well (50 genes, P < 1 × 10-15, phi coefficient = 0.413) (left), and also genes with H3K4me3 loss in Ras- and Raf-induced senescence overlapped well (380 genes, P < 1 × 10-15, phi coefficient = 0.559) (right). The overlap in H3K27me3 gain (Figure 10A) was again markedly rare, compared with the overlap in H3K4me3 gain (P = 2 × 10-9, log linear analysis) or with that in H3K4me3 loss (P < 1 × 10-15, log linear analysis).
- Citation: Fujimoto M, Mano Y, Anai M, Yamamoto S, Fukuyo M, Aburatani H, Kaneda A. Epigenetic alteration to activate Bmp2-Smad signaling in Raf-induced senescence. World J Biol Chem 2016; 7(1): 188-205
- URL: https://www.wjgnet.com/1949-8454/full/v7/i1/188.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v7.i1.188