Copyright
©2014 Baishideng Publishing Group Inc.
World J Biol Chem. May 26, 2014; 5(2): 141-160
Published online May 26, 2014. doi: 10.4331/wjbc.v5.i2.141
Published online May 26, 2014. doi: 10.4331/wjbc.v5.i2.141
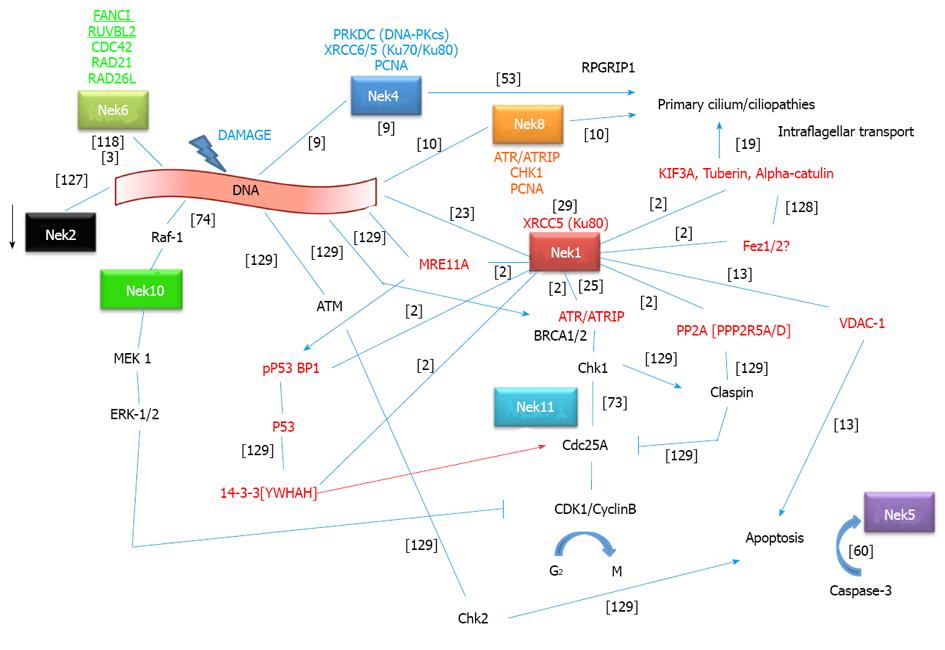
Figure 4 Nek1 interactome and crosstalk with other Neks and protein interactors in the context of the DNA damage response pathways.
Interactions between proteins are depicted as simple lines, activation is depicted as an arrow and inhibition as an arrow with a line as arrowhead. A red arrow for 14-3-3 means that it causes activation by the transport of CDC25 to the nucleus. Nek1 interacted with a specific 14-3-3 isoform called YWHAH[2] (gene symbols inside brackets correspond to the isoforms of those proteins which were described to interact with Nek1). Not necessarily the same specific 14-3-3 protein promotes the indicated functions. Rather, a family characteristic is intended to be assigned. Nek2 kinase activity is inhibited after DNA damage (arrow)[127]. The red protein names are those that have been identified to directly interact with Nek1 as identified by the yeast two-hybrid system[2] or other as indicated in the figure. Gene symbols above/under protein names represent other interactors of those proteins. Nek4 interactors have been identified by mass spectrometry[9]. As can be seen, all but three Neks (Nek3, 7 and 9) seem to be directly linked to the DNA damage response. Most strikingly, we can see a direct connection for Nek8, 4 and 1 between DDR and primary cilium function and ciliopathies. New connections to apoptosis have been recently pointed out for Nek1 and 5. References for interactions are depicted in brackets: Nek6[3,118]; Nek1[2,13,25]; Nek4[9,53]; Nek8[10]; Nek11[73]; Nek10[74]; Nek2[127]; Nek5[60]; KIF3A[19]; Fez1/2[128], various known interactions[129].
- Citation: Meirelles GV, Perez AM, de Souza EE, Basei FL, Papa PF, Melo Hanchuk TD, Cardoso VB, Kobarg J. “Stop Ne(c)king around”: How interactomics contributes to functionally characterize Nek family kinases. World J Biol Chem 2014; 5(2): 141-160
- URL: https://www.wjgnet.com/1949-8454/full/v5/i2/141.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v5.i2.141