Copyright
©2012 Baishideng Publishing Group Co.
World J Biol Chem. Jan 26, 2012; 3(1): 7-26
Published online Jan 26, 2012. doi: 10.4331/wjbc.v3.i1.7
Published online Jan 26, 2012. doi: 10.4331/wjbc.v3.i1.7
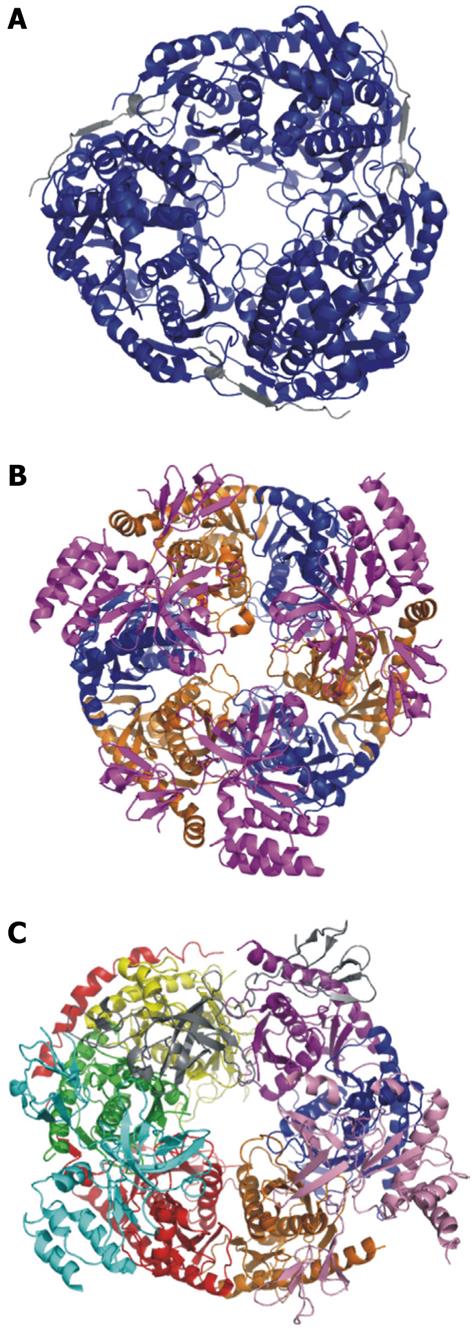
Figure 3 Crystal structures of the bacterial PNPase, archaeal exosome, and human exosome.
A: Escherichia coli PNPase. Pictured in blue is the polynucleotide nucleotidyltransferase domain. In grey is the associated RNAse E domain (RCSB # 3GCM). The S1 and KH domains are not pictured because they are not included in the crystallized complex; B: Archaeal exosome. In blue is aRrp42, in orange is aRrp41. These two proteins are the exonucleolytic core proteins. In magenta is the aRrp4 cap protein (RCSBID: 2BA0); C: Human exosome. In orange is Rrp41p, in Blue is Rrp42p, in yellow is Rrp43p, in red is Rrp45p, in green is Rrp46p, in purple is Mtr3p. These six proteins make up the scaffolding ring structure. Also pictured are the cap proteins; in pink is Rrp4, in cyan is Rrp40, and in grey is Csl4 (RCSBID: 2NN6). The S. cervisiae exosome has not been crystalized but the human and yeast exosomes have approximately 51% sequence identity[51,66,143].
- Citation: Bernstein J, Toth EA. Yeast nuclear RNA processing. World J Biol Chem 2012; 3(1): 7-26
- URL: https://www.wjgnet.com/1949-8454/full/v3/i1/7.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v3.i1.7