Copyright
©The Author(s) 2016.
World J Stem Cells. Oct 26, 2016; 8(10): 355-366
Published online Oct 26, 2016. doi: 10.4252/wjsc.v8.i10.355
Published online Oct 26, 2016. doi: 10.4252/wjsc.v8.i10.355
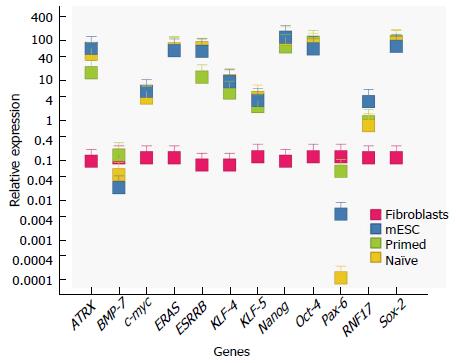
Figure 2 Differentially expressed genes in primed, naive and reprogrammed primed embryonic stem cells.
qRT-PCR of known master factor stem cell genes and candidate genes selected from our microarray analysis (Tables 3 and 4, Figure 4). Differences are measured in relative expression levels (to control fibroblasts). Results show that master factor genes such as Oct-4, Sox-2 and Nanog are all significantly higher than the control fibroblasts (red) in naive (blue), primed (green) and reprogrammed primed cells (yellow). Primers used are shown in Table 2. Esrrb, Atrx and Rnf-17 are all significantly upregulated in naïve ESCs and reprogrammed primed ESCs, relative to primed ESCs. Pax-6 and BMP-7 are significantly upregulated in primed ESCs. Expression levels were measured in established ESCs and primed ESCs after the 30th passage, in re-programmed primed ESCs two passages after transduction, and in fibroblasts two passages after primary cells were extracted. Error bars indicate SEM within cell populations (Tukey’s post hoc, P < 0.001; n = 5 replicates of independent cell lines). ESCs: Embryonic stem cells.
- Citation: Rossello RA, Pfenning A, Howard JT, Hochgeschwender U. Characterization and genetic manipulation of primed stem cells into a functional naïve state with ESRRB. World J Stem Cells 2016; 8(10): 355-366
- URL: https://www.wjgnet.com/1948-0210/full/v8/i10/355.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v8.i10.355