Copyright
©The Author(s) 2015.
World J Stem Cells. Jan 26, 2015; 7(1): 137-148
Published online Jan 26, 2015. doi: 10.4252/wjsc.v7.i1.137
Published online Jan 26, 2015. doi: 10.4252/wjsc.v7.i1.137
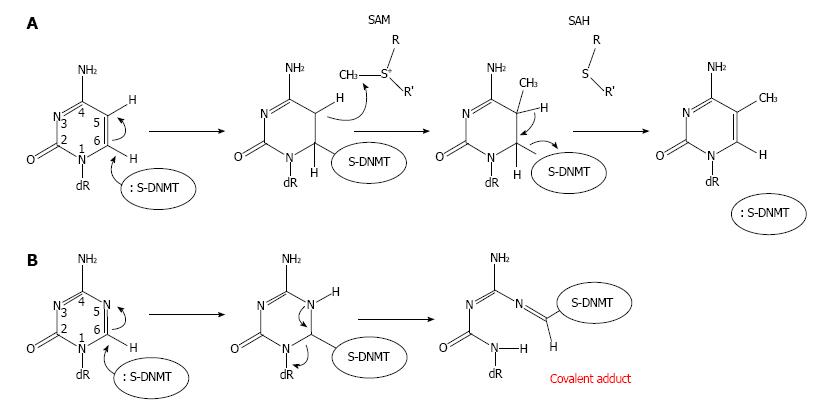
Figure 2 Mechanism of DNA methylation and its inhibition by 5-Aza-2’-deoxycytidine via covalent trapping.
A: Cytosine methylation by a DNMT is initiated by the formation of a covalent bond between the cytosine base and a cysteine residue at the active site of the methyltransferase. SAM then transfers a methyl group to the cytosine generating a methylcytosine. B: A DNMT is covalently trapped by the DNA-incorporated 5-Aza-2’-deoxycytidine (Aza-dC) adduct. The sulfhydryl group of the cysteine residue at the active site of DNMT forms a covalent bond with C6 of the Aza-dC in DNA. However, the active methyl group from SAM cannot be transferred to Aza-dC by DNMT. This covalent trapping mechanism then leads to a gradual loss of free DNMTs, because they are covalently linked with Aza-dC within DNA. Therefore, the loss of free DNMTs then precedes passive DNA demethylation. Thus cells expressing high levels of DNMTs will respond to Aza-dC at a higher extent compared with cells expressing low levels of DNMTs (see Figure 3). R: Methionine/cysteine; R’: Adenosine; Dr: Deoxyribose-5-phosphate; DNMT: DNA methyltransferases.
- Citation: Wongtrakoongate P. Epigenetic therapy of cancer stem and progenitor cells by targeting DNA methylation machineries. World J Stem Cells 2015; 7(1): 137-148
- URL: https://www.wjgnet.com/1948-0210/full/v7/i1/137.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v7.i1.137