Copyright
©The Author(s) 2021.
World J Stem Cells. Jun 26, 2021; 13(6): 485-502
Published online Jun 26, 2021. doi: 10.4252/wjsc.v13.i6.485
Published online Jun 26, 2021. doi: 10.4252/wjsc.v13.i6.485
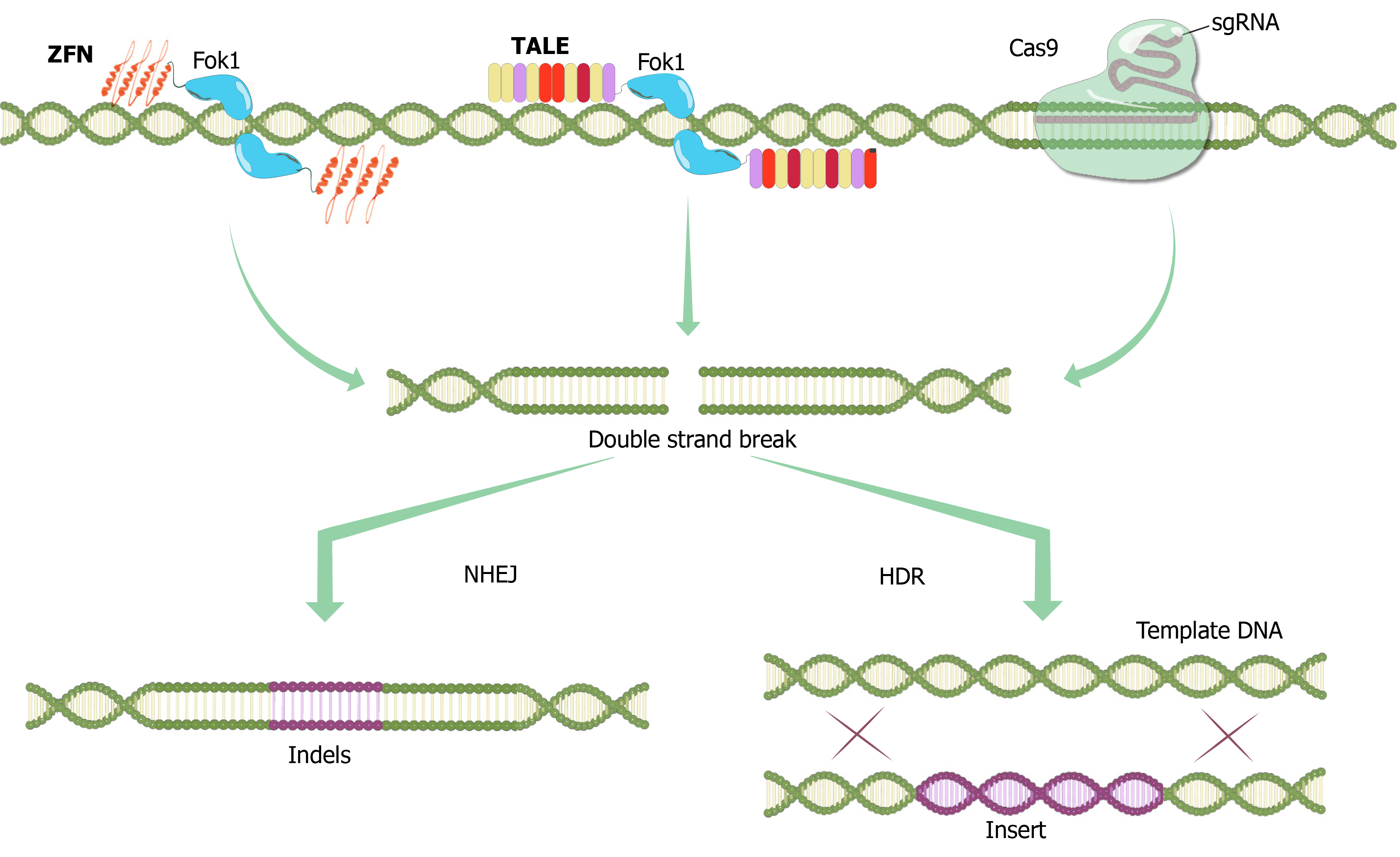
Figure 1 Programmable nucleases.
Zinc finger nucleases contain a specially designed N-terminal DNA-binding domain and a C-terminal endonuclease domain (bacterial Fok1 endonucleases) that dimerize to form a double-strand break (DSB) in DNA[111]. DNA-binding domain is a sequence of multiple zinc finger domains, each of which recognizes a specific three base pair sequence of DNA. Then, four to six zinc finger regions recognize specific 12-18 base pair nucleotide sequences. Transcription activator-like effector nucleases contain a C-terminal Fok1 endonuclease with an N-terminal DNA binding site. The DNA binding domain consists of highly conserved 33-35 amino acids with a variation at the 12th and 13th positions that ensure specificity against the target sequence. CRISPR/Cas9system; crRNA-tracrRNA (sgRNA) and Cas9 protein bind to DNA by forming a complex. The Cas9 enzyme has two nuclease sites. These are the HNH region (which creates a break in the complementary chain) and the RuvC region (which creates a break in the non-complementary chain). In the presence of the Protospacer adjacent motif (PAM) sequence, the sgRNA-Cas9 complex binds and cuts DNA generating DSBs. ZFN: Zinc Finger Nuclease; TALE: Transcription Activator-Like Effector; sgRNA: single guide RNA; NHEJ: Non-homologous end joining; HDR: Homology directed repair.
- Citation: Eksi YE, Sanlioglu AD, Akkaya B, Ozturk BE, Sanlioglu S. Genome engineering and disease modeling via programmable nucleases for insulin gene therapy: Promises of CRISPR/Cas9 technology . World J Stem Cells 2021; 13(6): 485-502
- URL: https://www.wjgnet.com/1948-0210/full/v13/i6/485.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i6.485