Copyright
©The Author(s) 2021.
World J Gastroenterol. Jul 28, 2021; 27(28): 4555-4581
Published online Jul 28, 2021. doi: 10.3748/wjg.v27.i28.4555
Published online Jul 28, 2021. doi: 10.3748/wjg.v27.i28.4555
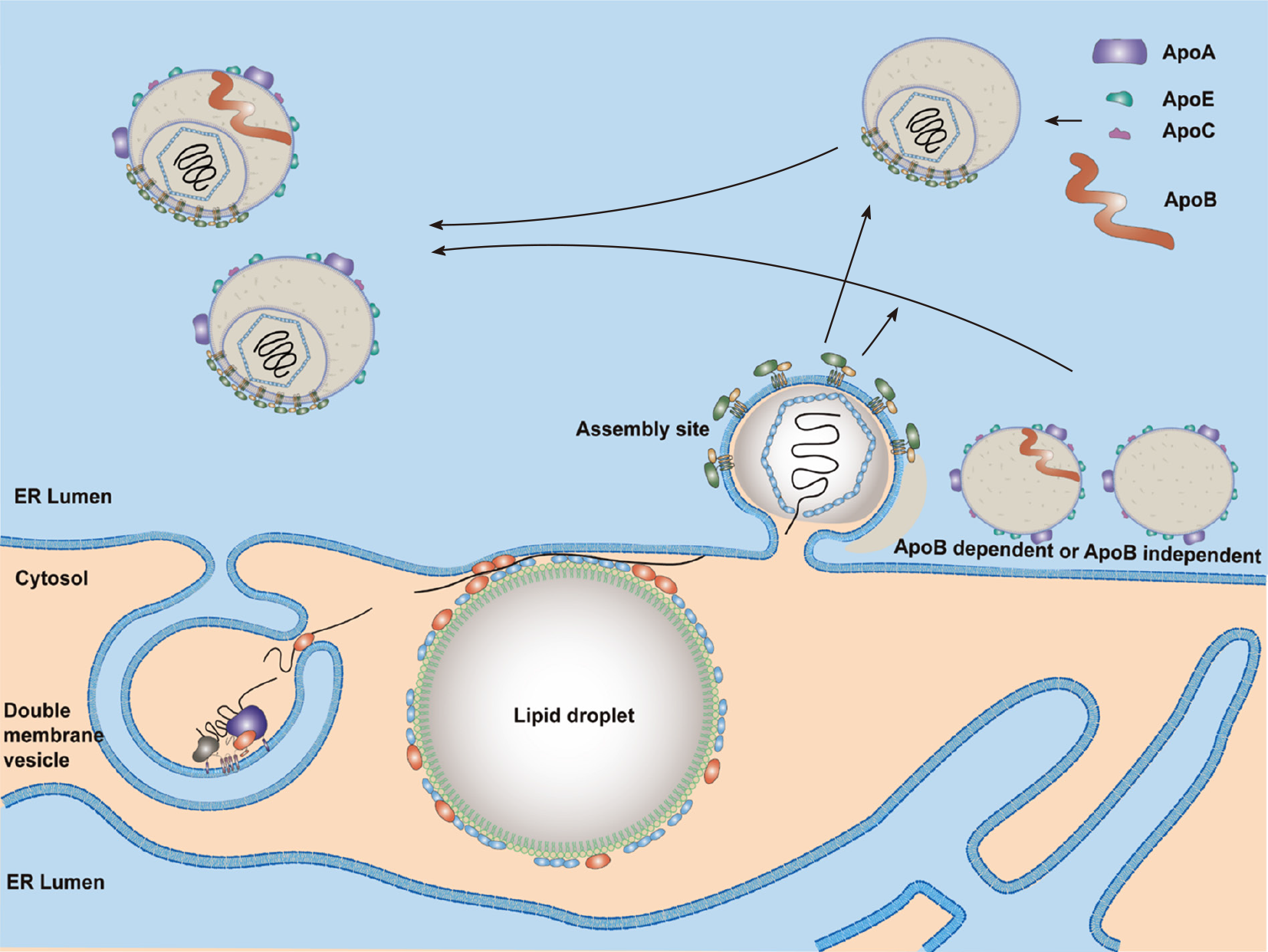
Figure 4 A proposed model for hepatitis C virus assembly.
Lipid droplets (LDs) are surrounded by endoplasmic reticulum (ER)[321]. LDs with a hepatitis C virus (HCV) core and NS5A proteins are close to replication sites [double membrane vesicle (DMV)] and assembly sites. HCV genomic RNA synthesized by the replication complex (NS3–NS5B proteins) in the DMVs will be transferred by NS5A and NS3-4A proteins and encapsidated by the core proteins to form the nucleocapsid. Then, the HCV nucleocapsid will interact with glycoproteins E1/E2 in the assembly sites and bud into the ER lumen. Both Apo-B-dependent and -independent mechanisms are possibly involved in HCV particle assembly. One model shows the production of a fused form of HCV with very-low-density lipoproteins. Another model shows the budding of HCV particles with several apolipoproteins but not Apo-B. Nascent viral particles may be further lipidated by luminal lipoproteins and incorporated with exchangeable apolipoproteins. ER: Endoplasmic reticulum.
- Citation: Li HC, Yang CH, Lo SY. Cellular factors involved in the hepatitis C virus life cycle. World J Gastroenterol 2021; 27(28): 4555-4581
- URL: https://www.wjgnet.com/1007-9327/full/v27/i28/4555.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i28.4555