Copyright
©The Author(s) 2021.
World J Gastroenterol. Jul 28, 2021; 27(28): 4555-4581
Published online Jul 28, 2021. doi: 10.3748/wjg.v27.i28.4555
Published online Jul 28, 2021. doi: 10.3748/wjg.v27.i28.4555
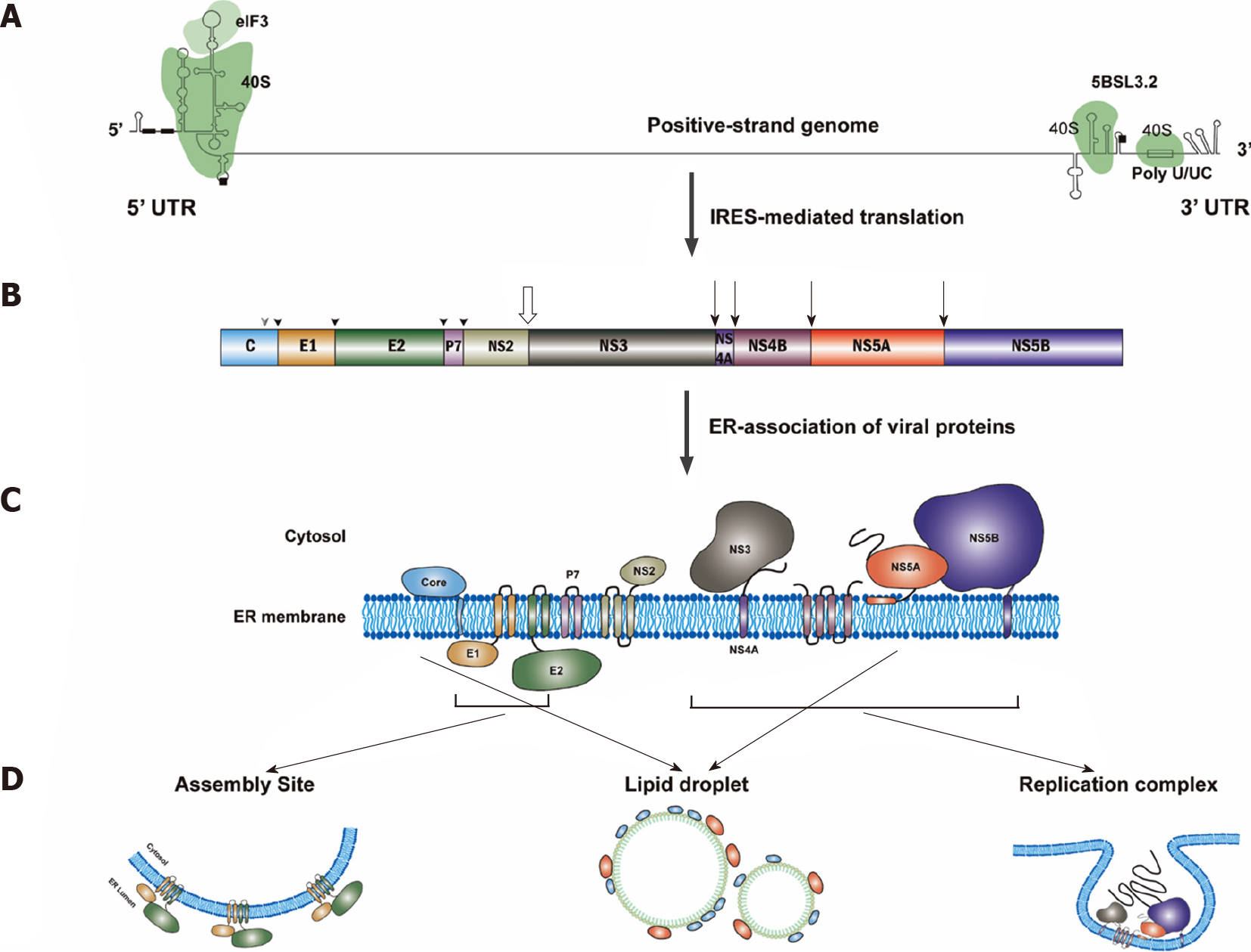
Figure 3 Hepatitis C virus protein translation.
A: Translation of hepatitis C virus (HCV) genomic RNA is regulated by the internal ribosome entry site in the 5’-untranslated region (5’UTR) along with a short segment of the core gene sequence, the cis-acting replication element in the NS5B coding region and by the entire 3’UTR. Binding sites for eIF3 and 40S were marked. The start and stop codons for protein translation are marked by black squares, while two recognition sites on the 5’UTR for miR122 are marked by black rectangles; B: Polyprotein is co- and post-translationally cleaved by host or viral proteases to yield the structural proteins (core, E1 and E2) and the nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B proteins). Core, E1 and E2 are processed by cellular signal peptidase (filled arrowhead). Mature core protein will be generated after further cleavage by signal peptide peptidase (empty arrowhead)[317]. The NS2/NS3 junction site is cleaved by the NS2–NS3 auto-protease[318] (empty arrow), and the remaining nonstructural proteins are processed by the NS3/4A proteinase[319] (filled arrow); C: All of the HCV proteins are associated with endoplasmic reticulum directly or indirectly[320]; D: Then, NS3, NS4A, NS4B, NS5A and NS5B proteins will form the replication complex. Core and NS5A proteins will be transferred to lipid droplets, while E1 and E2 proteins will stay in the assembly sites. IRES: Internal ribosome entry site; 5’UTR: 5’-untranslated region.
- Citation: Li HC, Yang CH, Lo SY. Cellular factors involved in the hepatitis C virus life cycle. World J Gastroenterol 2021; 27(28): 4555-4581
- URL: https://www.wjgnet.com/1007-9327/full/v27/i28/4555.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i28.4555