Copyright
©The Author(s) 2019.
World J Gastroenterol. Apr 21, 2019; 25(15): 1783-1796
Published online Apr 21, 2019. doi: 10.3748/wjg.v25.i15.1783
Published online Apr 21, 2019. doi: 10.3748/wjg.v25.i15.1783
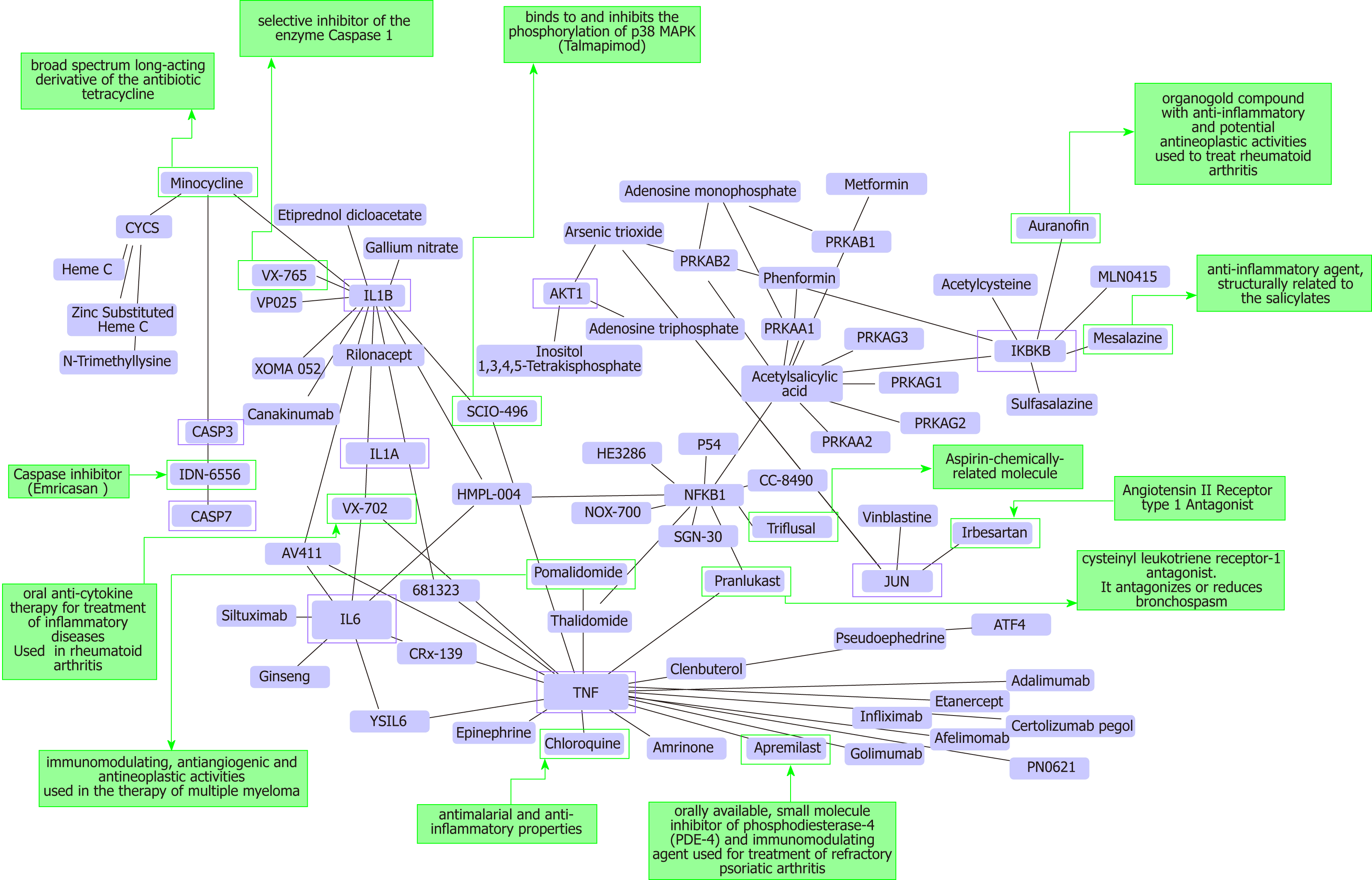
Figure 3 Protein-chemical interactions and potential repurposing drugs to target nonalcoholic steatohepatitis.
We generated a protein-chemical interaction network by mapping the significant genes/proteins that are represented in the nonalcoholic fatty liver disease-Kyoto Encyclopedia of Genes and Genomes pathway to chemicals/drugs that are annotated in the Comparative Toxicogenomics Database. The 149 genes (seeds) from our analysis were mapped to the corresponding molecular interaction database; full list of seed genes is listed in Table 1. This analysis generated a huge network composed of approximately 2000 nodes. Current figure shows chemical-drug-interactions specifically focused on selected genes/proteins of potential interest, including members of the caspase family (CASP3 and CASP7), interleukins (IL1B, IL1A, and IL6), tumor necrosis factor α (TNF-α), NFKB1 (Nuclear factor kappa B subunit 1) and IKBKB (inhibitor of nuclear factor kappa B kinase subunit beta), JUN (Jun proto-oncogene, AP-1 transcription factor subunit), AKT1 (AKT serine/threonine kinase 1). In green charts we summarized information on current use and known action of selected drugs. Interaction network was predicted by the Networkanalyst resource available at https://http://www.networkanalyst.ca/faces/home.xhtml. The network is shown as a Cytoscape graph.
- Citation: Sookoian S, Pirola CJ. Repurposing drugs to target nonalcoholic steatohepatitis. World J Gastroenterol 2019; 25(15): 1783-1796
- URL: https://www.wjgnet.com/1007-9327/full/v25/i15/1783.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i15.1783