Copyright
©The Author(s) 2016.
World J Gastroenterol. Sep 21, 2016; 22(35): 7938-7950
Published online Sep 21, 2016. doi: 10.3748/wjg.v22.i35.7938
Published online Sep 21, 2016. doi: 10.3748/wjg.v22.i35.7938
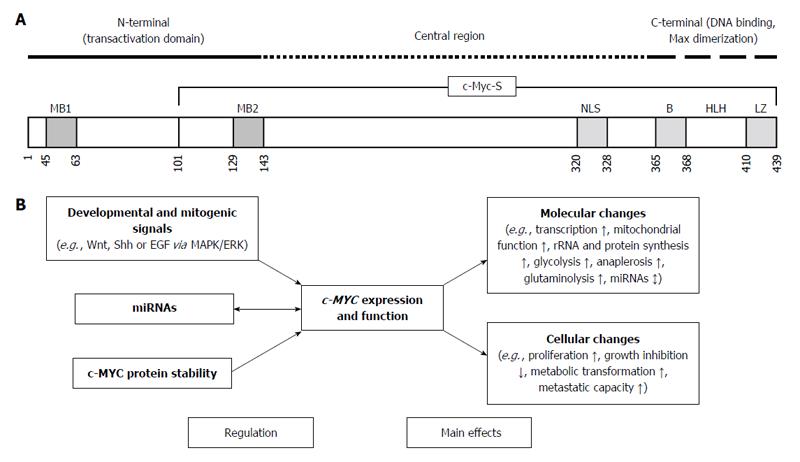
Figure 1 Schematic representation of the structure, regulation and main effects of c-MYC.
A: The c-MYC protein consists of three domains: N-terminal, Central region, and C-terminal. The Central region and the C-terminal domain of c-MYC are responsible for protein-protein interactions that result in transcriptional repression by c-MYC. The C-terminal domain contains a basic (B) helix-loop-helix (HLH)-leucine zipper (LZ) motif that is necessary for interaction with different proteins (such as Max), and physiological recognition of DNA target sequences[7,58]; B: Developmental and mitogenic signals tightly regulate c-MYC gene expression both in normal (nontransformed) and in transformed cells via the MAPK/ERK pathway. MicroRNAs display a dual-faced role in the c-MYC regulatory network; both as regulators and as targets of c-MYC. The stability of the c-MYC protein also represents a particularly effective mechanism of gene regulation. c-Myc-S: Truncated c-Myc protein; MB1 and MB2: Evolutionarily conserved Myc Box sequences; NLS: Nuclear localization signal; Shh: Sonic hedgehog; EGF: Epidermal growth factor; MAPK: Mitogen-activated protein kinase; ERK: Extracellular signal-regulated kinase.
- Citation: Sipos F, Firneisz G, Műzes G. Therapeutic aspects of c-MYC signaling in inflammatory and cancerous colonic diseases. World J Gastroenterol 2016; 22(35): 7938-7950
- URL: https://www.wjgnet.com/1007-9327/full/v22/i35/7938.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i35.7938