Copyright
©The Author(s) 2015.
World J Gastroenterol. Jan 21, 2015; 21(3): 711-725
Published online Jan 21, 2015. doi: 10.3748/wjg.v21.i3.711
Published online Jan 21, 2015. doi: 10.3748/wjg.v21.i3.711
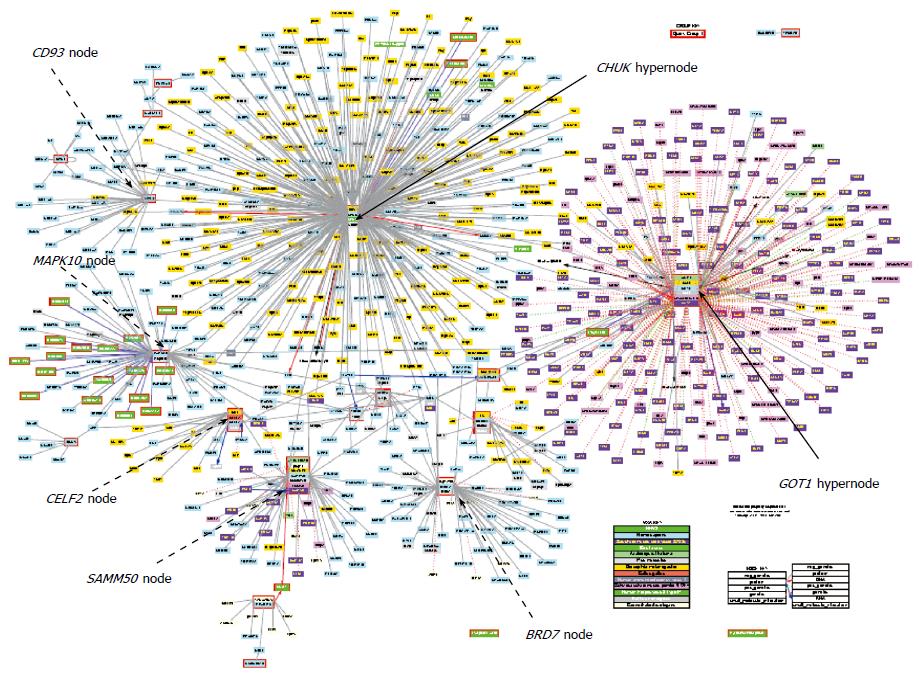
Figure 2 Visualization of biomolecular interactions among associated loci with serum levels of alanine-aminotransferase and aspartate-aminotransferase in published genome-wide association studies.
Prediction was based on the Cognoscente program, freely available at the web-based submission portal: http://vanburenlab.tamhsc.edu/cognoscente.html. The interaction network image shows 828 nodes with different levels of complexity; black arrows indicate the major nodes. Additional interconnectivity nodes of importance (highlighted in black dashed arrows) are SAMM50, CD93 (highly connected with CHUK), MAPK10, CELF2 and BRD7. Prediction by Cognoscente supports multiple organisms in the same query, as well as gene-gene, gene-protein, protein-RNA and protein-DNA interactions, and multi-molecule queries[77]. The input list was based on the gene list presented in Table 4, while the graph depicts known interactions the query list members.
- Citation: Sookoian S, Pirola CJ. Liver enzymes, metabolomics and genome-wide association studies: From systems biology to the personalized medicine. World J Gastroenterol 2015; 21(3): 711-725
- URL: https://www.wjgnet.com/1007-9327/full/v21/i3/711.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i3.711