Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Jul 7, 2014; 20(25): 7993-8004
Published online Jul 7, 2014. doi: 10.3748/wjg.v20.i25.7993
Published online Jul 7, 2014. doi: 10.3748/wjg.v20.i25.7993
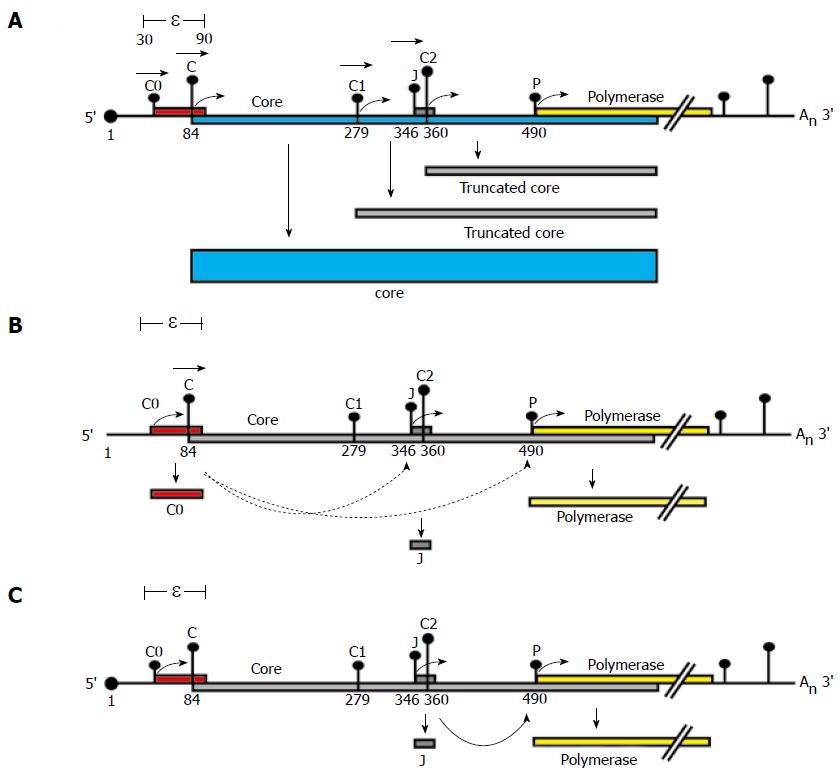
Figure 4 Proposed model for P protein synthesis.
A schematic representation of a pgRNA transcript and its coding regions. The pgRNA transcript encodes the core (blue) and polymerase (yellow) genes. The pgRNA transcript start site is numbered 1. The pgRNA contains the ε structure and a repeated sequence of 117 bases in both its 5′ and 3′ ends. Vertical bars with dots represent the initiation codons within the pgRNA labelled C0, C, C1, J, C2 and P. The height of each bar represents the match to the "ideal" initiation context (Kozak’s consensus)[15]. Filled boxes represent ORFs encoded by each initiation codon, (A)n denotes the polyA tail and the black dot at the 5′ end denotes the cap structure. The proposed model for P protein synthesis extends a scanning model. A: Leaky scanning. Ribosomes would scan from the capped 5′ end of the pgRNA, ribosomes would predominantly scan past C0 (in a poor initiation context) and initiate at the C AUG codon (which has an optimal initiation context) synthesising the core. However, some ribosomes may continue to leaky scan (represented by horizontal arrowheads) past the C AUG initiating at distal AUGs (e.g. making truncated core proteins from C1 or C2); B: Role of C0. The C0 initiation codon is the first in the pgRNA, some ribosomes would initiate here, when they terminate they could re-initiate at P or J (represented by lower curved arrows). Those that initiate at P would translate the polymerase; C: Role of J. Ribosomes which bypass C and C1 AUG codons by leaky scanning would then initiate translation at the J AUG codon (optimal initiation context), translating the short J ORF effectively bypassing the C2 AUG codon. Ribosomes which translated the J ORF may then reinitiate at the P AUG codon, translating the polymerase (curved arrow).
- Citation: Chen A, T-Thienprasert NP, Brown CM. Prospects for inhibiting the post-transcriptional regulation of gene expression in hepatitis B virus. World J Gastroenterol 2014; 20(25): 7993-8004
- URL: https://www.wjgnet.com/1007-9327/full/v20/i25/7993.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i25.7993