Copyright
©2007 Baishideng Publishing Group Co.
World J Gastroenterol. Jan 7, 2007; 13(1): 48-64
Published online Jan 7, 2007. doi: 10.3748/wjg.v13.i1.48
Published online Jan 7, 2007. doi: 10.3748/wjg.v13.i1.48
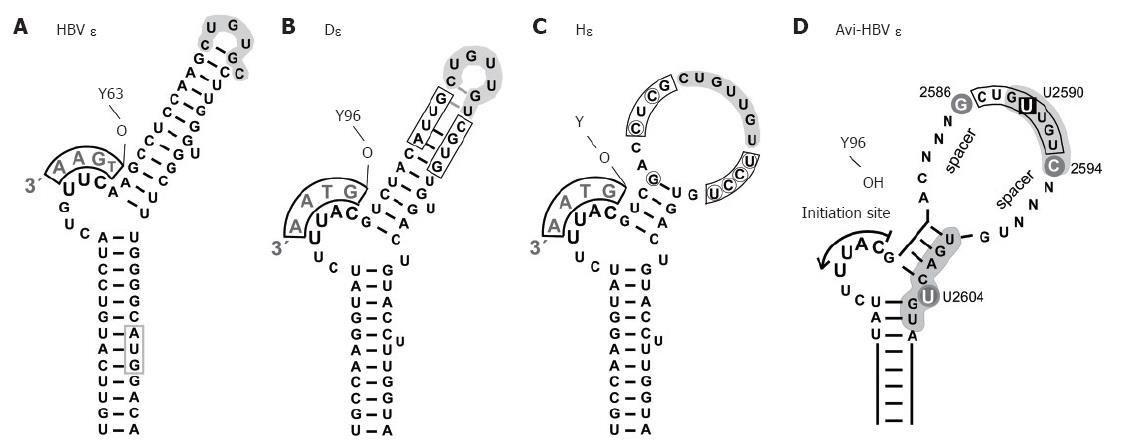
Figure 4 Secondary structures of hepadnaviral ε signals.
A: HBV ε. The entire hairpin, including in the upper stem region, is stably base-paired; formation of a stable tri-loop, as indicated, is confirmed by direct NMR analysis[68]. The conventional annotation for the loop sequence is high-lighted by grey shading. The bulge-templated DNA oligonucleotide and the priming Y63 are indicated; B: DHBV ε. The overall 2D structure is similar to that of HBV ε, including a largely base-paired upper stem[69], as confirmed by preliminary NMR data. The boxed positions were randomized in a SELEX approach, and P binding individuals were selected from the corresponding RNA library[71]; C: HHBV ε (Hε). Hε shows substantial sequence variation to D: (encircled nt), leading to a largely open upper stem structure; D: Generalized secondary structure of an avihepadnaviral ε signal. The scheme summarizes major determinants for productive interaction with P protein[71]. Grey background indicates nt positions that are probably in contact with protein. The bulge and its immediate vicinity, particularly the tip of the lower stem, are essential for P binding whereas the loop appears critically involved in the transition to a productive initiation complex[109]. U2590 > C or U2604 > G mutations abrogate, or strongly reduce, P binding whereas G2586 and C2594 do not affect binding but are important for priming. The major role of the nt termed N may be to provide a proper spacing to the bulge element; their sequence is not important as long as formation of highly stable base-pairing is prevented.
- Citation: Beck J, Nassal M. Hepatitis B virus replication. World J Gastroenterol 2007; 13(1): 48-64
- URL: https://www.wjgnet.com/1007-9327/full/v13/i1/48.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i1.48