Copyright
©The Author(s) 2023.
World J Gastroenterol. Jun 14, 2023; 29(22): 3385-3399
Published online Jun 14, 2023. doi: 10.3748/wjg.v29.i22.3385
Published online Jun 14, 2023. doi: 10.3748/wjg.v29.i22.3385
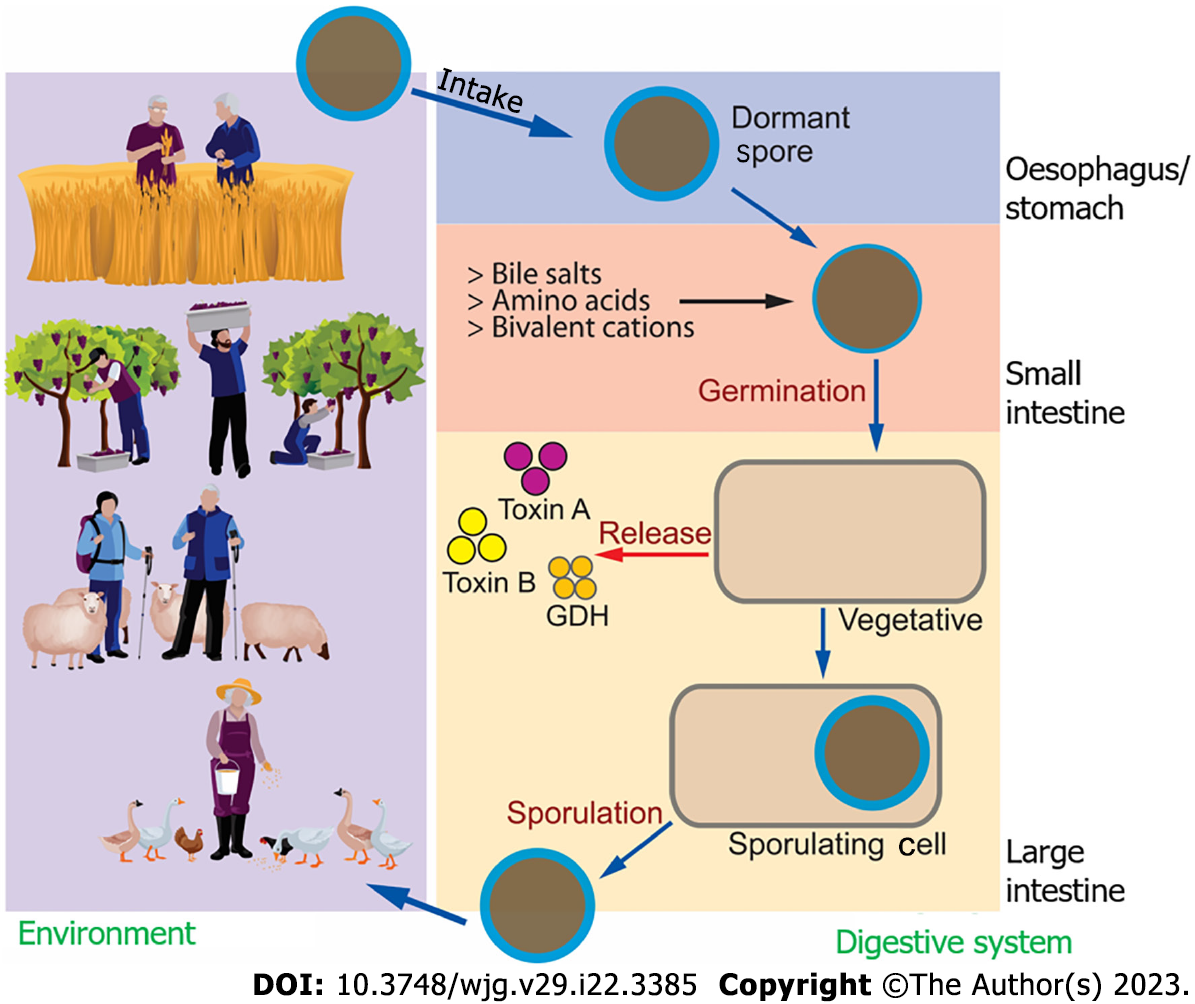
Figure 1 Clostridioidesdifficile life cycle.
Clostridioides difficile (C. difficile) can be found in soil and is transmitted to humans (as well as animals) through different daily activities. Besides hospital-acquired C. difficile infections, community-acquired C. difficile infections are also known, with agriculture being one of the main sources, from the spore stage into the human digestive system. Once stable, C. difficile may repopulate and release toxins that can affect intestinal health. C. difficile may go back to the environment and repeat the life cycle. GDH: Glutamate dehydrogenase.
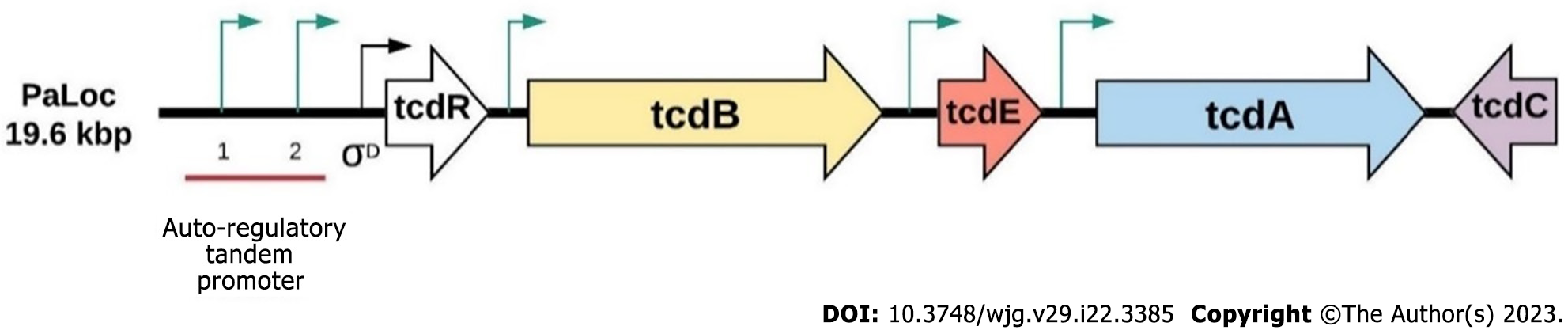
Figure 2 Pathogenicity loci of toxicogenic Clostridioidesdifficile.
The 19.6 kbp long pathogenicity loci in toxicogenic Clostridioides difficile, including tcdC, tcdA, tcdE, tcdB and tcdR coding genes. tcdR codes for an RNA polymerase sigma factor that controls the expression of the tcdB and tcdA genes and possibly tcdE. The transcription of tcdR can be regulated by three promoters. One is under the control of σD, and the other two are autoregulatory tandem promoters. Figure adopted and modified from Isidro et al[19].
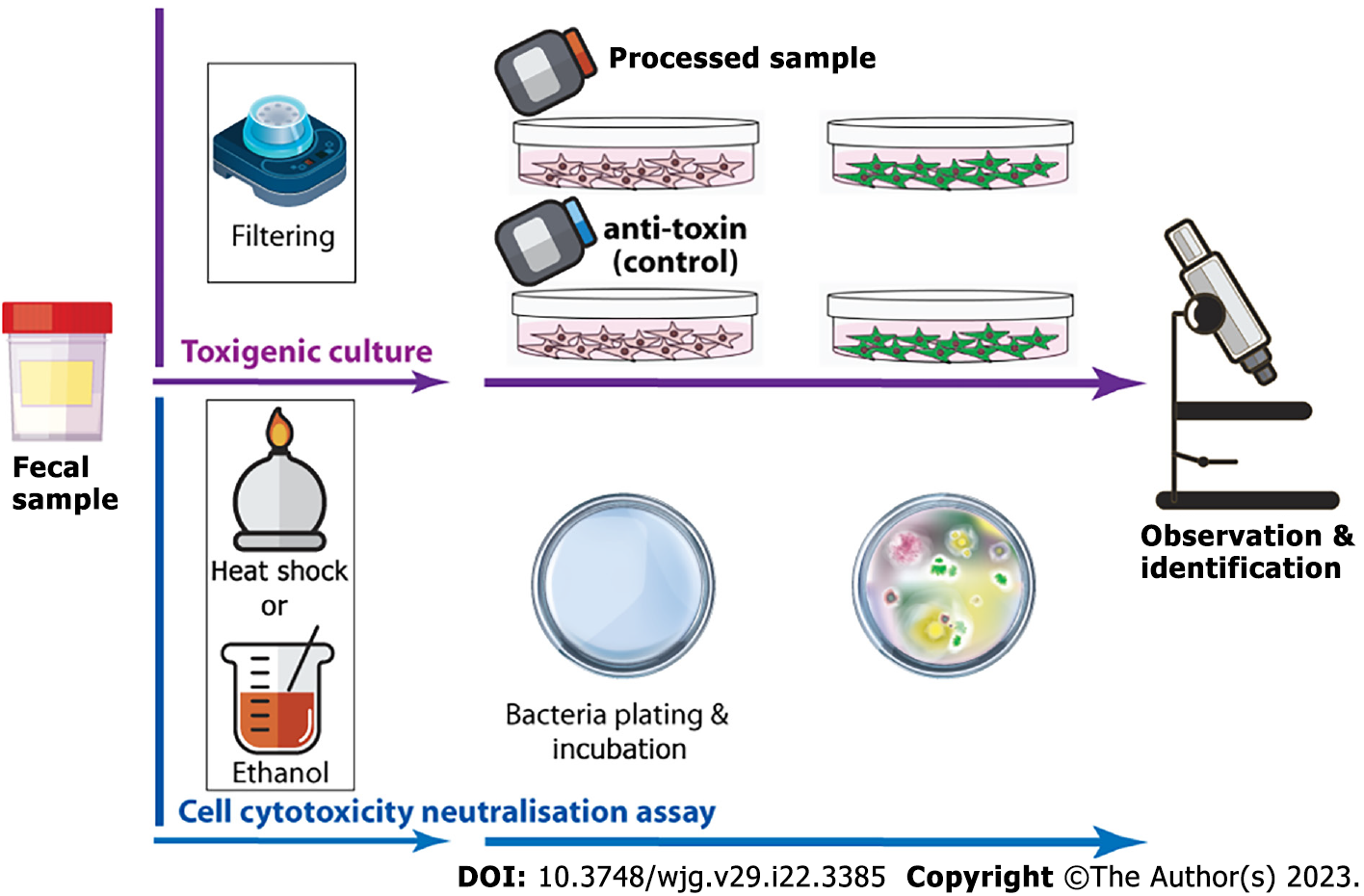
Figure 3 Idealised summary of toxigenic culture and cell cytotoxicity neutralisation assay for Clostridioidesdifficile infection identification.
In toxigenic culture, the faecal sample is first exposed to heat shock or alcohol shock to kill Clostridioides difficile (C. difficile) and other microorganisms, and heat/alcohol resistant spores of C. difficile survive. The sample is then plated on selective agar and incubated anaerobically at 37 °C for at least 48 h to observe and confirm the colonisation of C. difficile. For the cell cytotoxicity neutralisation assay, the faecal sample is filtered and added to a toxin-sensitive cell line with/without antiserum. After 24-48 h of incubation, the cytopathological effect is then examined microscopically.
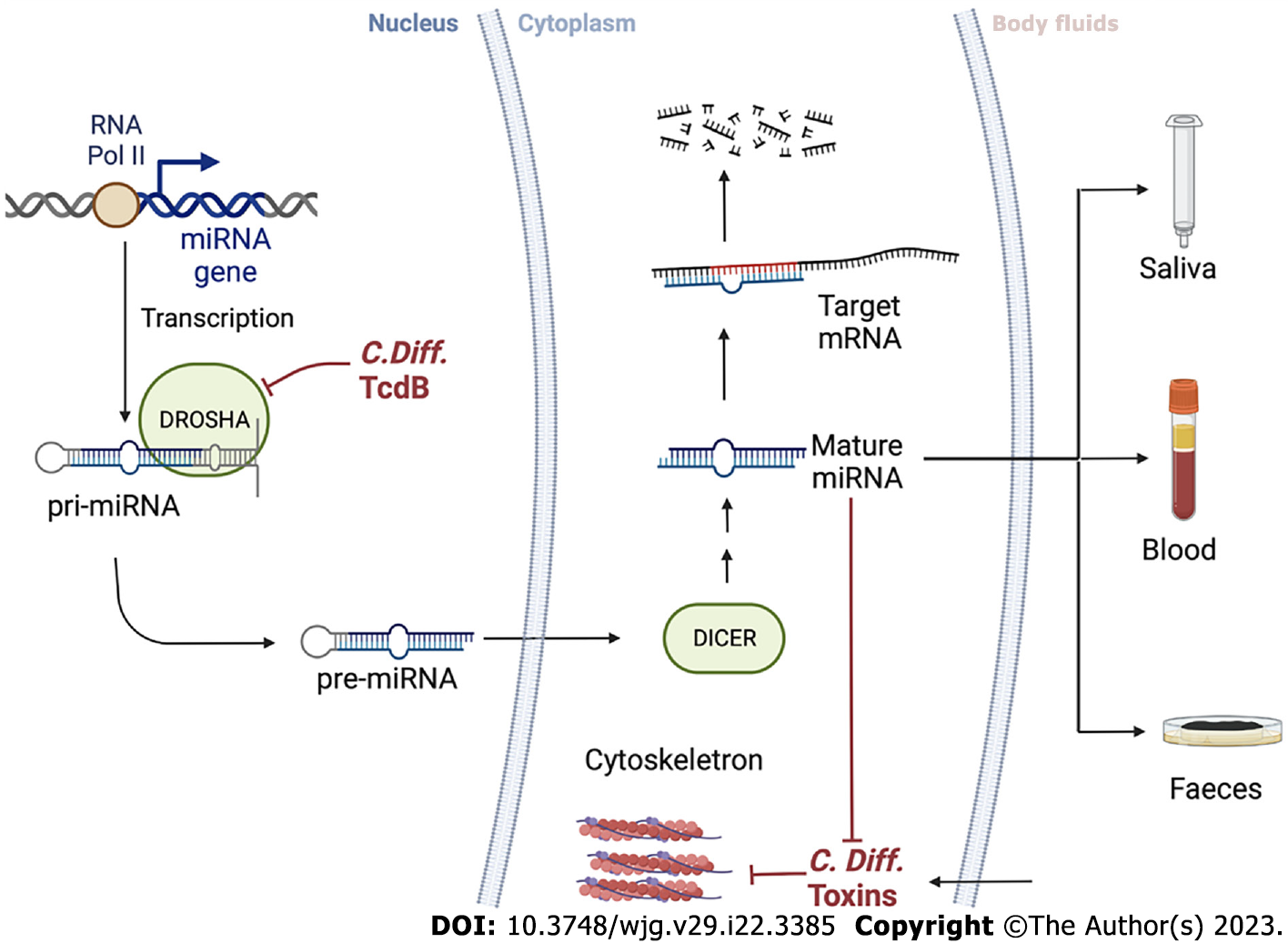
Figure 4 Clostridioidesdifficile and microRNAs cross-interact inside the cells.
microRNA (miRNA) is a small single-stranded RNA that plays a key role in gene expression regulation. It starts in the cell nucleus, where primary miRNAs are produced by RNA Pol II through miRNA transcription from DNA sequences or miRNA genes. Precursor miRNA, around 60-110 nucleotides long transcripts with a shorter stem-loop structure, are produced from primary miRNAs by RNase type III (DROSHA) enzyme with several maturation processes and is then transported to the cytoplasm via DICER. Toxin B from Clostridioides difficile may interfere with DROSHA function. A fraction of miRNA binding specific sequences (2-8 nucleotides long) can pair with miRNA response elements in the 3’ untranslated region of the target mRNA, causing translational repression and degradation of the target mRNA. Thus, miRNA expression patterns can indicate non-physiological events such as Clostridioides difficile infection. By detecting changes in miRNA patterns in body fluids, such as saliva, blood, and stool, the screening accuracy can be improved. miRNA: microRNA; pri-miRNA: Primary miRNA; pre-miRNA: Precursor miRNA; C. difficile: Clostridioides difficile.
- Citation: Bocchetti M, Ferraro MG, Melisi F, Grisolia P, Scrima M, Cossu AM, Yau TO. Overview of current detection methods and microRNA potential in Clostridioides difficile infection screening. World J Gastroenterol 2023; 29(22): 3385-3399
- URL: https://www.wjgnet.com/1007-9327/full/v29/i22/3385.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i22.3385