Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Aug 7, 2014; 20(29): 9775-9827
Published online Aug 7, 2014. doi: 10.3748/wjg.v20.i29.9775
Published online Aug 7, 2014. doi: 10.3748/wjg.v20.i29.9775
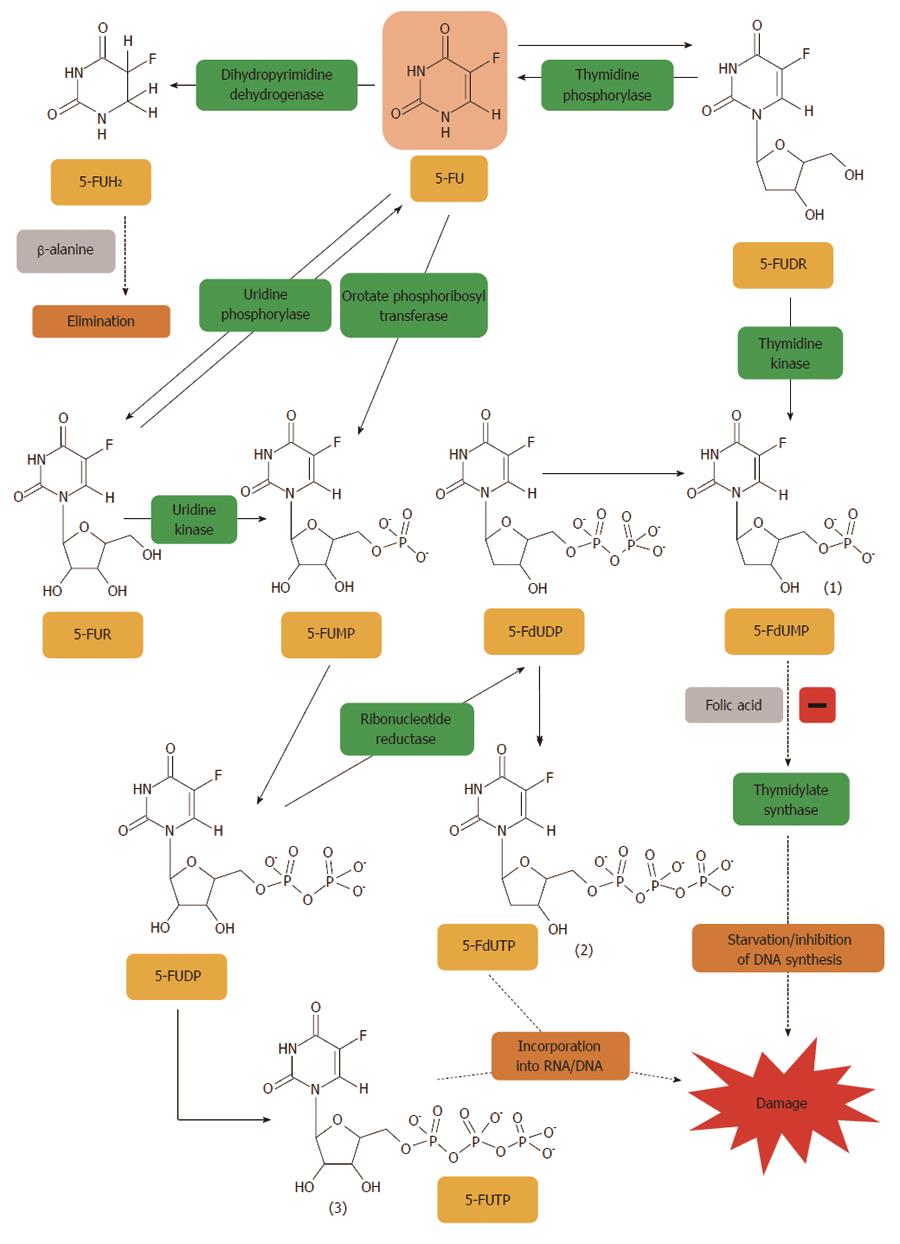
Figure 1 5-fluorouracil is converted to three major active metabolites.
(1) fluorodeoxyuridine monophosphate (FdUMP); (2) fluorodeoxyuridine triphosphate (FdUTP); and (3) fluorouridine triphosphate (FUTP). The main mechanism of 5-fluorouracil (5-FU) activation is conversion to fluorouridine monophosphate (FUMP) either directly by orotate phosphoribosyl transferase (OPRT), or indirectly via fluorouridine (FUR) through the sequential action of uridine phosphorylase and uridine kinase. FUMP is then phosphorylated to fluorouridine diphosphate (FUDP), which can be either further phosphorylated to the active metabolite fluorouridine triphosphate (FUTP), or converted to fluorodeoxyuridine diphosphate (FdUDP) by ribonucleotide reductase. In turn, FdUDP can either be phosphorylated or dephosphorylated to generate the active metabolites FdUTP and FdUMP, respectively. An alternative activation pathway involves the thymidine phosphorylase catalyzed conversion of 5-FU to 5-fluoro-2’-deoxyuridine (5-FUDR), which is then phosphorylated by thymidine kinase to the thymidylate synthase inhibitor, FdUMP. Dihydropyrimidine dehydrogenase (DPD)-mediated conversion of 5-FU to dihydrofluorouracil (DHFU) is the rate-limiting step of 5-FU catabolism in normal and tumour cells[401].
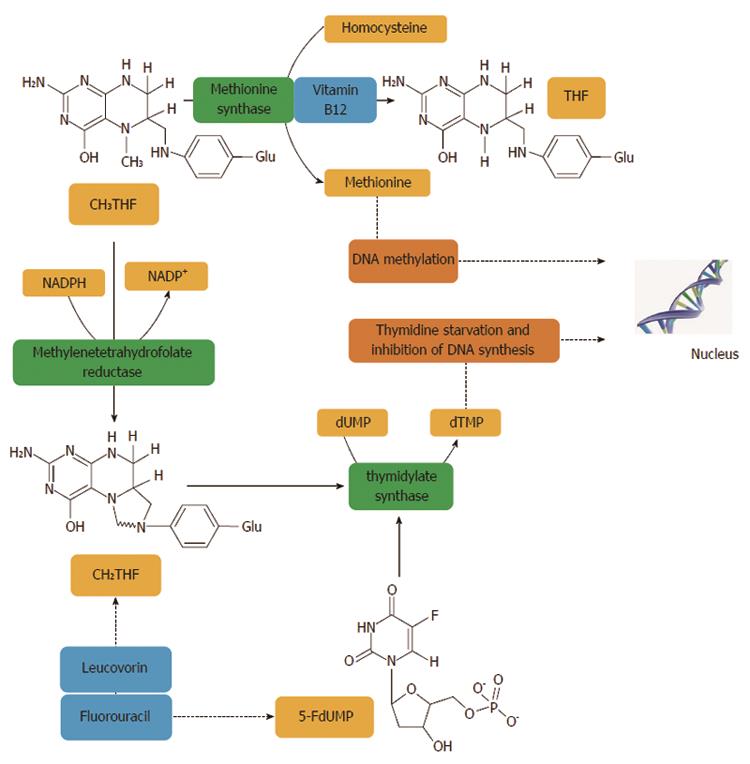
Figure 2 Methylentetrahydrofolate reductase plays an important role in the action of 5-fluorouracil, an inhibitor of thymidylate synthase.
Methylentetrahydrofolate reductase (MTHFR) catalyses a unidirectional reaction that lowers the levels of 5,10-methylenetetrahydrofolate (CH2THF) by increasing levels of 5-methyltetrahydrofolate (CH3THF) which is used for biological methylation. Other factors, such as vitamin B12 and homocysteine, are involved in biological methylation processes. The addition of folinic acid (leucovorin) to 5-FU improves the response rates and survival of CRC patients. Thymidylate synthase (TS) catalyses the reductive methylation of deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP) with the reduced folate, CH2THF, as the methyl donor. This reaction provides the sole de novo source of thymidylate, which is necessary for DNA replication and repair. TS contains a nucleotide-binding site and a binding site for CH2THF. The 5-FU metabolite, FdUMP, binds to the nucleotide-binding site of TS, forming a stable ternary complex with the enzyme and CH2THF which blocks binding of the normal substrate dUMP, thereby inhibiting dTMP synthesis. Inhibition of thymidylate synthesis causes disruption of nucleotide levels that results in DNA damage[402].
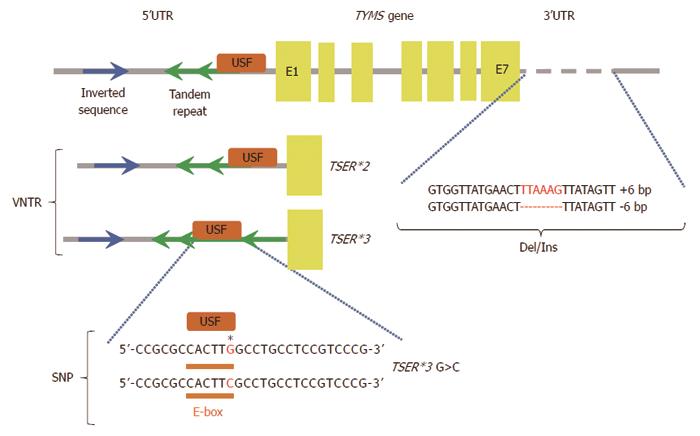
Figure 3 Some of the described polymorphisms affect inter-individual differences in patient sensitivity to 5-fluorouracil treatment.
Polymorphisms in the thymidylate synthase gene (TYMS gene), 5’ and 3’ untranslated regions (5’UTR and 3’UTR), exons (E1-E7), binding site for upstream stimulating factor (USF), variable number tandem repeats (VNTR), single nucleotide polymorphism (SNP), deletion/insertion polymorphism (Del/Ins), two-tandem repeats (TSER*2), three-tandem repeats (TSER*3), TSER*3 G>C (single nucleotide polymorphism of TSER*3). Regulation of TYMS gene expression. TSER polymorphism (TS 2R/3R repeat) is a tandem repeat upstream of the TYMS translational start site containing either double (2R) or triple (3R) repeats of 28-bp sequences. These tandem repeats regulate transcription and translation of TYMS. Additional functional variants of the TYMS gene have been identified and TSER 2R/3R repeat is now studied together with a G to C SNP within the second repeat of the 3R allele. TSER 3RC/3RC genotype causes lower transcriptional activity of TYMS, comparable with the TS 2R/2R genotype. TS 1494del6bp is another functional variant of the TYMS gene and has been shown to decrease RNA stability and therefore influence TS mRNA and TS protein expression in vitro[52].
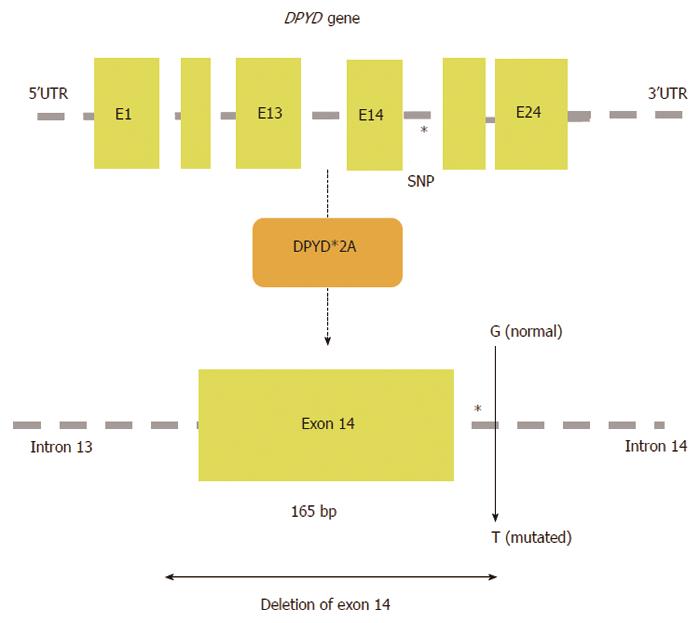
Figure 4 A schematic map of the human DPYD gene is shown with the location of SNP DPYD*2A (IVS14+1G>A); exon 14 is skipped as a result of the G>A translocation at intron 14.
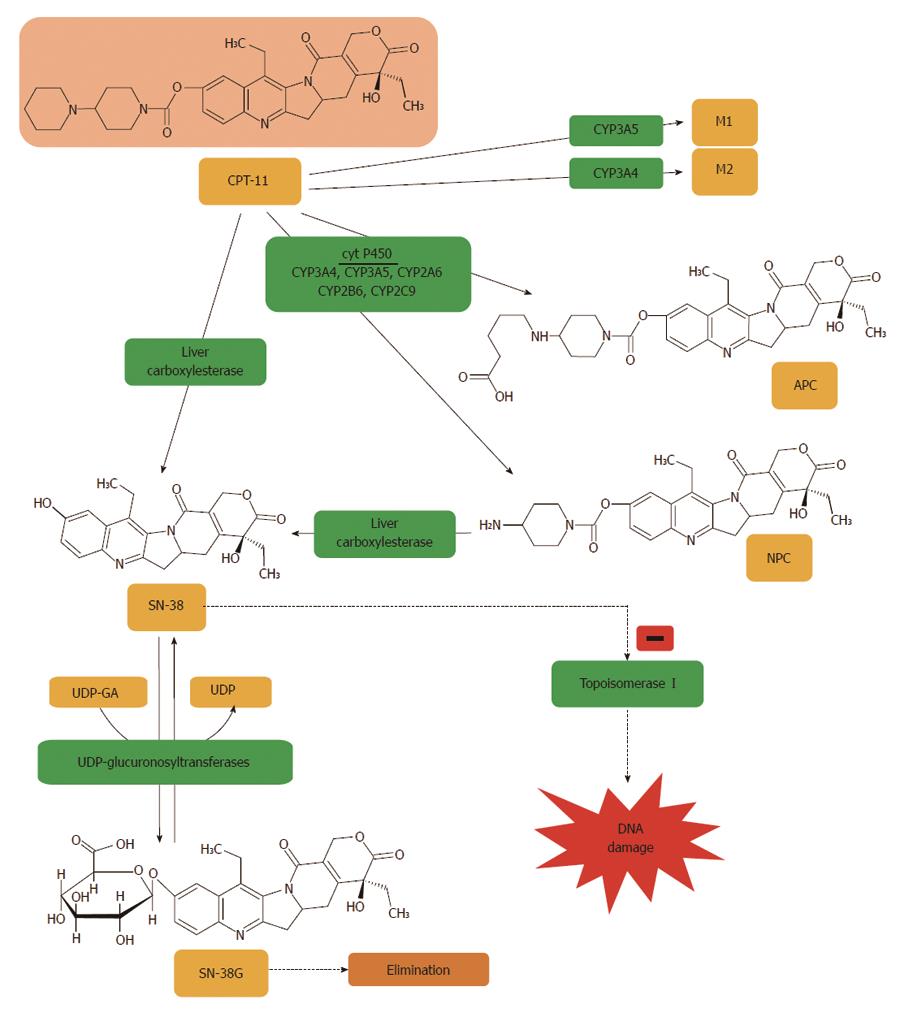
Figure 5 Irinotecan is metabolized to APC or NPC and potential other intermediate metabolites (M1, M2) via a cytochrome P450 mediated process.
Neither 7-ethyl-10-[4-N-(5-aminopentanoic acid)-1-piperidino]carbonyloxycamptothecin (APC) nor 7-ethyl-10-(4-amino-1-piperidino)carbonyloxycamptothecin (NPC) contribute directly to irinotecan activity in vivo. NPC is further converted to 7-ethyl-10-hydroxy-camptothecin (SN-38) by carboxylesterase. All irinotecan metabolites are pH sensitive, thus are at risk of transforming from inactive to active products, and vice versa. SN-38 is subsequently conjugated predominantly by the enzyme UDP-glucuronosyltransferase 1A1 (UGT1A1) to form a glucuronide metabolite (SN-38G)[403].
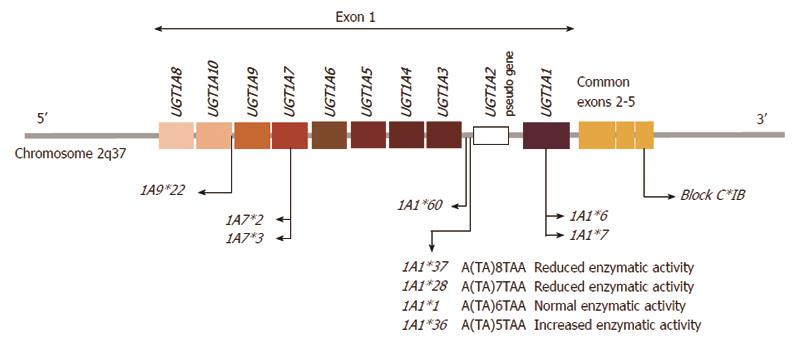
Figure 6 Graphic representation of the human UGT1A gene locus encoding the UGT1A enzymes and the major UGT1A1, 1A7 and 1A9 polymorphisms that are responsible for glucuronidation of SN-38.
Individual first exons are positioned at the 5’ end of the chromosome and common exons 2-5 at the 3’ end. Individual exon 1 sequences are combined with exons 2-5 sequence, which is present in every UDP-glycosyltransferase 1A1 (UGT1A1) transcript, the intervening sequence of the primary transcript is eliminated by splicing[404]. The promoter variant, UGT1A1*28, *36 and *37 results from a TA insertion/deletion in the (TA)6TAA element of the UGT1A1 promoter region. This alteration leads to decreased/increased gene expression[184].
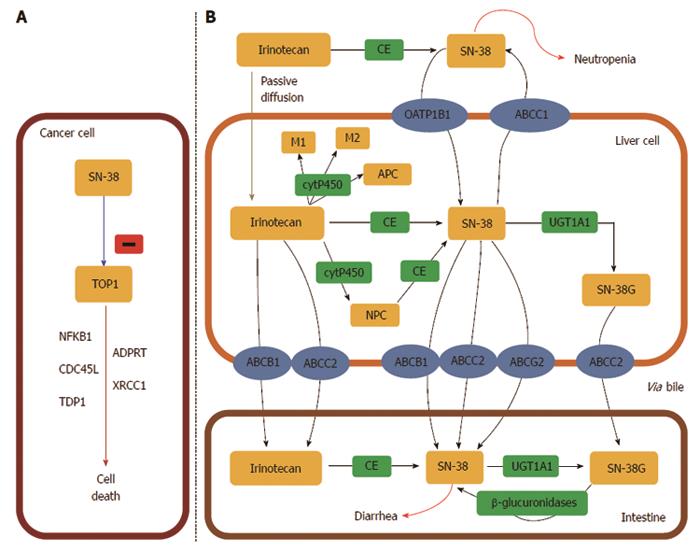
Figure 7 UDP-glycosyltransferase 1 family.
A: The active metabolite of irinotecan, SN-38, is a DNA topoisomerase I (TOP1) inhibitor which leads to cancer cell death. TOP1 is a nuclear enzyme required in replication, responsible for unwinding DNA and preventing lethal strand breaks. SN-38 is cytotoxic and destabilizes the TOP1-DNA covalent complex formed in colorectal cancer cells. SN-38 causes irreversible double strand breaks which lead to S phase arrest followed by cell death. To do so, SN-38 attaches to the complexes and blocks future replication forks preventing repair of double strand breaks[405]; B: Irinotecan uptake and transport into the liver is facilitated by: OATP1B1 (SLCO1B1), ABCB1, MRP1 (ABCC1), MRP2 (ABCC2), and MXR (ABCG2). Specifically, ABCB1 is present on the bile membrane and is responsible for the secretion of irinotecan and its metabolites into the liver[406]. Irinotecan is metabolized in the liver and converted to SN-38, the active metabolite and TOP1 inhibitor, by carboxylesterases (CE) mediated hydrolysis. SN-38 is then glucoronized to SN-38 glucuronic acid (SN-38G) and detoxified in the liver via conjugation by the UGT1A family, which releases SN-38G into the intestines for elimination[407]. Approximately 70% of SN-38 becomes SN-38G, which has 1/100 of the antitumour activity and is virtually inactive. In the intestinal lumen, bacterial β-glucuronidases can reverse the reaction and transform inactive SN-38G back into the active form SN-38. This factor contributes to varied toxicity, specifically dose limiting diarrhoea[198].
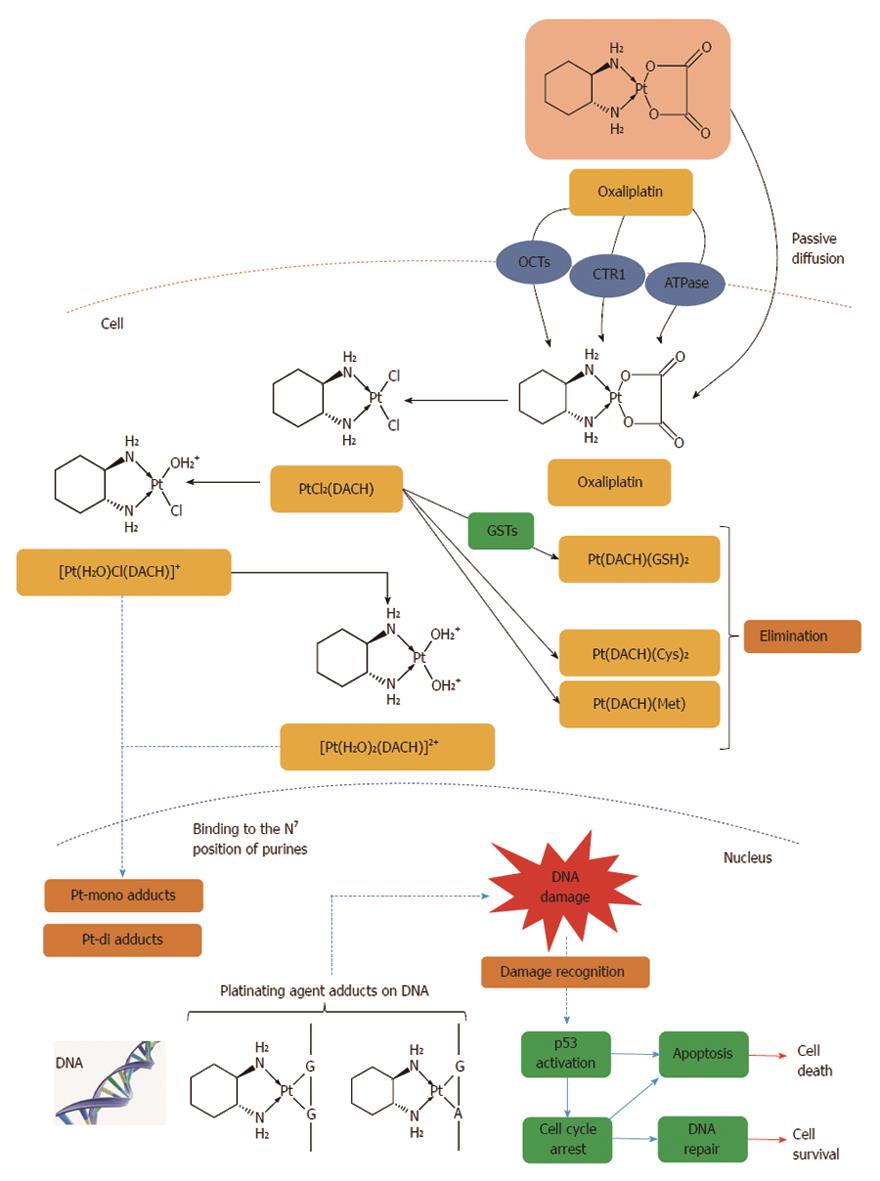
Figure 8 Intracellular drug accumulation.
The free fraction of oxaliplatin is biotransformed non-enzymatically and subsequently forms complexes with chloride, glutathione (GSH), methionine (Met) and cysteine (Cys). Oxaliplatin undergoes non-enzymatic conversion in physiologic solutions to active derivatives via displacement of the labile oxalate ligand. Several transient reactive species are formed, including monoaquo DACH (1,2-diaminocyclohexane) platinum [Pt(H2O)Cl(DACH)]+ and diaquo DACH platinum [Pt(H2O)2(DACH)]2+, which covalently bind with macromolecules. There is no evidence of cytochrome P450-mediated metabolism in vitro. The major route of platinum elimination is renal excretion. The main mechanism of action is mediated through the formation of DNA adducts which is thought to be related to the anti-tumour effects of oxaliplatin. An important factor is the induction of apoptosis by the primary DNA-Pt lesions, which is possibly enhanced by the contribution of targets other than DNA. Several influx and efflux transporters such as organic cation transporters (OCTs) 1, 2 and 3 (SLC22A1, SLC22A2 and SLC22A3), copper efflux transporters (CTRs), P-type ATPases, ATP7A and ATP7B have been identified, which may play an important role in determining tumour sensitivity and/or resistance to oxaliplatin[408].
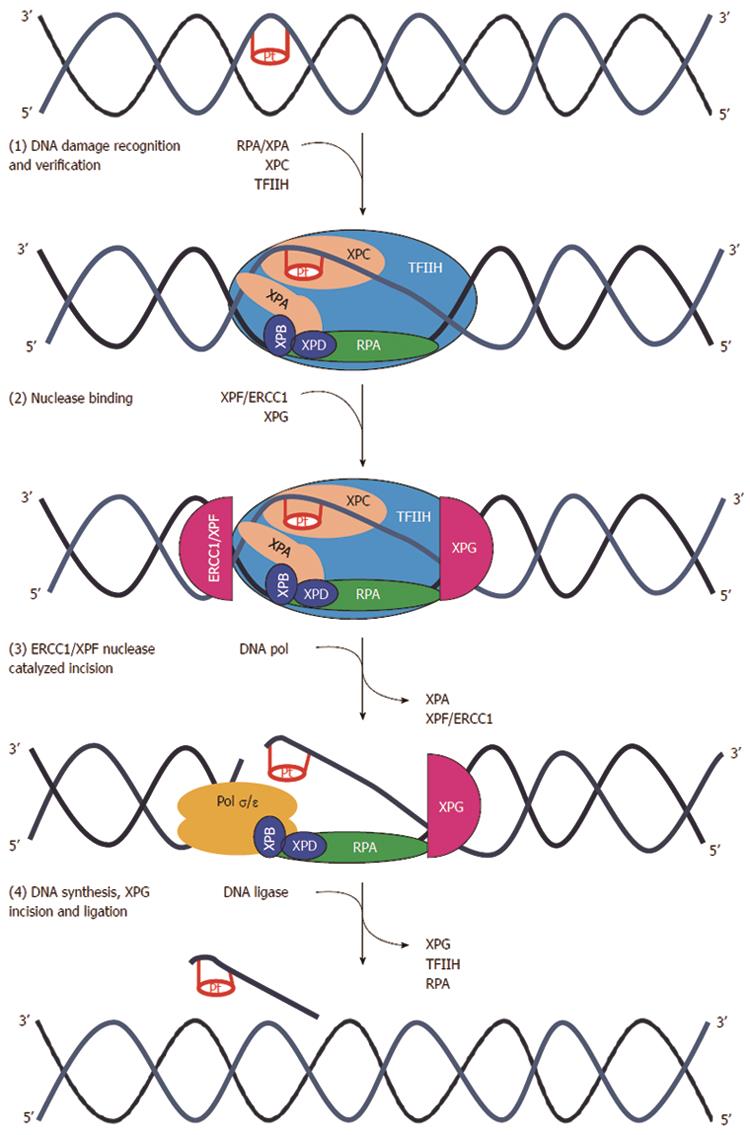
Figure 9 Nucleotide excision repair pathway.
(1) DNA damage formed by platinum agents leads to DNA double helix distortion. Several distinct complexes are involved in sequential steps than can be summarized as DNA damage recognition (XPCHR23B), damage demarcation, and verification (TFIIH), assembly of a preincision complex (RPA and XPA) and helix unwinding (XPB and XPD); (2) Endonuclease recruitment with dual incision of the damaged strand on the 5’ side (ERCC1-XPF heterodimers) and 3’ side (XPG) followed by the removal of the excised oligomer; (3) DNA repair synthesis to fill in the resulting gap; and (4) ligation. ERCC1: Excision repair cross-complementation group 1; Pol σ/ε : Polymerase σ/ε; RFC: Replication factor C; TFIIH: Transcription factor II H; XP (A,B,C,D,F,G): Xeroderma pigmentosum complementation group (A,B,C,D,F,G)[340].
- Citation: Panczyk M. Pharmacogenetics research on chemotherapy resistance in colorectal cancer over the last 20 years. World J Gastroenterol 2014; 20(29): 9775-9827
- URL: https://www.wjgnet.com/1007-9327/full/v20/i29/9775.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i29.9775