Published online Oct 28, 2020. doi: 10.37126/aige.v1.i2.33
Peer-review started: September 21, 2020
First decision: September 25, 2020
Revised: October 1, 2020
Accepted: October 13, 2020
Article in press: October 13, 2020
Published online: October 28, 2020
Processing time: 36 Days and 8.4 Hours
Wireless capsule endoscopy (WCE) enables physicians to examine the gastrointestinal tract by transmitting images wirelessly from a disposable capsule to a data recorder. Although WCE is the least invasive endoscopy technique for diagnosing gastrointestinal disorders, interpreting a WCE study requires significant time effort and training. Analysis of images by artificial intelligence, through advances such as machine or deep learning, has been increasingly applied to medical imaging. There has been substantial interest in using deep learning to detect various gastrointestinal disorders based on WCE images. This article discusses basic knowledge of deep learning, applications of deep learning in WCE, and the implementation of deep learning model in a clinical setting. We anticipate continued research investigating the use of deep learning in interpreting WCE studies to generate predictive algorithms and aid in the diagnosis of gastrointestinal disorders.
Core Tip: Wireless capsule endoscopy is the least invasive endoscopy technique for investigating the gastrointestinal tract. However, it takes a significant amount of time for interpreting the results. Deep learning has been increasingly applied to interpret capsule endoscopy images. We have summarized deep learning’s framework, various characteristics in published literature, and application in the clinical setting.
- Citation: Atsawarungruangkit A, Elfanagely Y, Asombang AW, Rupawala A, Rich HG. Understanding deep learning in capsule endoscopy: Can artificial intelligence enhance clinical practice? Artif Intell Gastrointest Endosc 2020; 1(2): 33-43
- URL: https://www.wjgnet.com/2689-7164/full/v1/i2/33.htm
- DOI: https://dx.doi.org/10.37126/aige.v1.i2.33
Since 1868, endoscopy has been constantly evolving and improving to assess the lumen and mucosa of the gastrointestinal tract, including the esophagus, stomach, colon, and parts of the small bowel[1]. Despite its utility, endoscopic examination of the small intestine is limited by its length and distance from accessible orifices[2-4]. This limitation is a factor that contributed to the development of wireless capsule endoscopy (WCE).
Developed in the mid-1990s, WCE utilizes an ingestible miniature camera that can directly view the esophagus, stomach, entire small intestine, and colon without pain, sedation, or air insufflation[2,5-7]. An important clinical application of WCE is the evaluation of gastrointestinal bleeding after a high quality bidirectional conventional endoscopy and colonoscopy does not identify a source of bleeding[5]. A typical WCE study lasts 8 to 12 h and generates 50000-100000 images. Reviewing that quantity of images requires significant time effort and training. Additionally, abnormalities in the gastrointestinal tract may be present in only one or two frames of the video which may be missed due to oversight[2]. An automatic computer-aided diagnosis system may aid and support physicians in their analysis of images captured by WCE.
Artificial intelligence (AI), an aspect of computer-aided design, has been rapidly expanding and permeating in academia and industry[8]. AI involves computer programs that perform functions associated with human intelligence[9,10]. Specific features of AI include computer learning and problem solving. AI was first described as the development of computer systems to perform tasks that require human intelligence, which can include decision making and speech recognition[11]. Many techniques of AI have been proposed to facilitate the recognition and prediction of patterns[12].
Machine learning (ML) is an application of AI that provides systems with the ability to automatically learn and improve from experience without explicit programming[13]. ML can recognize patterns from datasets to create algorithms and make predictions[10,12]. A tremendous breakthrough in ML has been the development of deep neural networks (also known as deep learning)[13]. Deep learning consists of massive multilayer networks of artificial neurons that can automatically discover useful features. To put it simply, deep learning can extract more patterns from high dimensional data[5,12]. Several deep learning models have been reported in the literature and are differentiated by their application[12]. Convolutional neural network (CNN), a type of deep learning, is highly effective at performing image analysis[8,13,14]. Given CNN’s utility in image analysis, applications for CNN have extended into the medical field, including gastroenterology[8,14]. The main drawback of deep learning is a long training time. Advances in graphic processing units, however, have drastically reduced the training time of deep learning from days or weeks to hours or days[15].
ML and CNN have been increasingly explored and applied to diagnostic images found in radiology, pathology, and dermatology[15-18]. Likewise, ML and CNN have utility in endoscopy and WCE through image-based interpretation without alteration of the existing procedures[8,11]. Current applications of ML and CNN in gastroenterology include polyp detection, esophageal cancer diagnosis, and ulcer detection through image-based interpretation from WCE. WCE is among the top interests of AI researchers in gastroenterology.
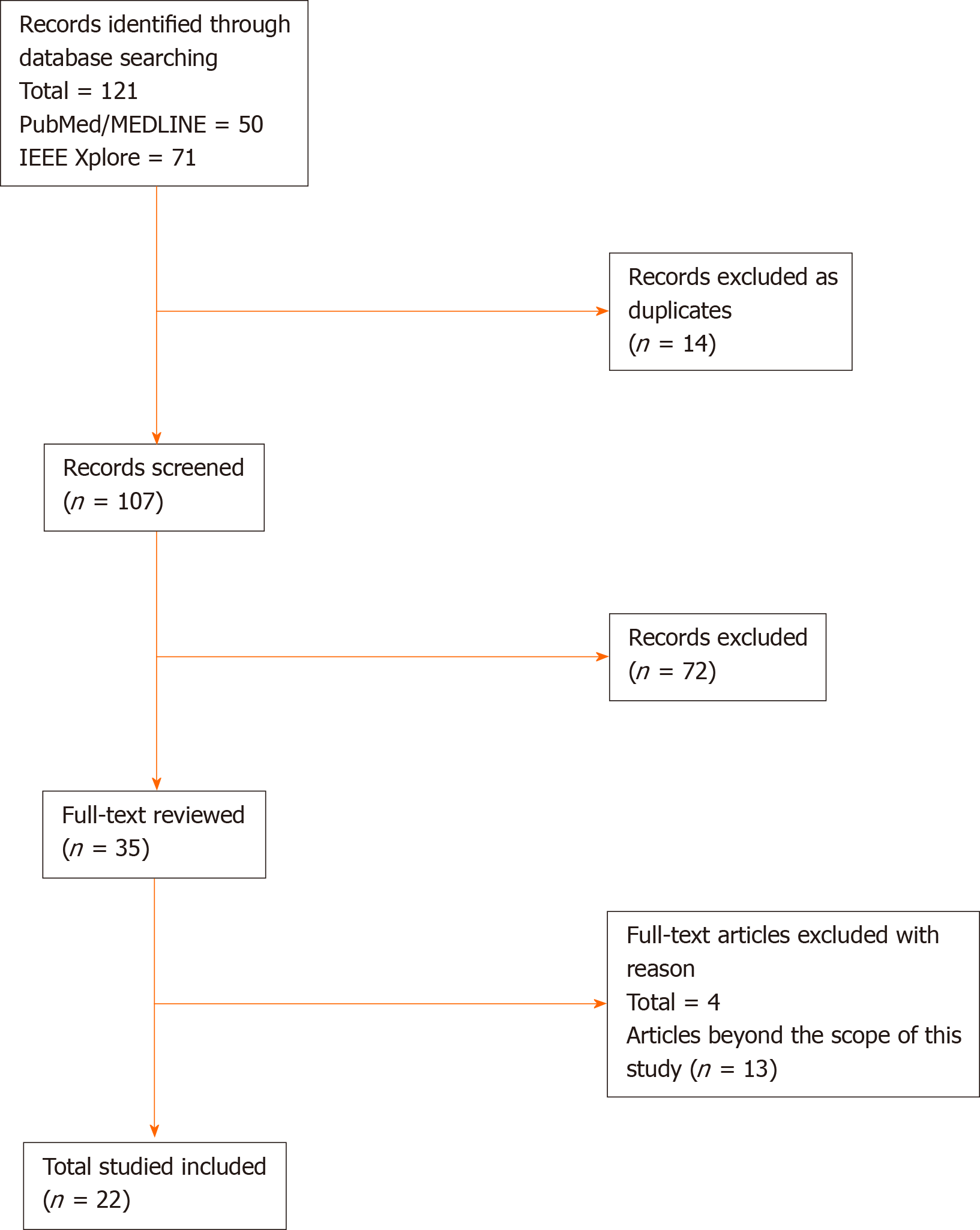
We conducted a literature review on December 15, 2019 and updated it on March 31, 2020 on PubMed/MEDLINE database and IEEE Xplore digital library. The search phrase used for query data in PubMed/MEDLINE database was ("Capsule Endoscopy") AND ("Deep Learning" OR "Neural Network" OR "Neural Networks"). Similarly, the search phrase used for query data in IEEE Xplore digital library was ("All Metadata":"Capsule Endoscopy") AND (("All Metadata":"Deep Learning") OR ("All Metadata":"Neural Network") OR ("All Metadata":"Neural Networks")). As presented in Figure 1, we found 50 records in PubMed/MEDLINE database and 71 records in IEEE Xplore digital library. After removing 14 duplicate records, the total number of distinct records were 107.
Only articles written in English language or available in English translation were considered. Conference abstracts, review articles, magazine articles, and unpublished studies were excluded to ensure quality. At this stage, two authors (AA and YE) independently reviewed whether the studies met the above inclusion criteria based on the title and abstract. Then, the articles that passed the initial screening were independently reviewed again based on the full-text articles to locate all included studies within a predefined scope of this article.
The most common indication for using WCE is the evaluation of small intestinal bleeding. WCE has also be used to diagnose other small intestinal disorders, such as celiac disease, Crohn’s disease, polyps, and tumors, for the evaluation of esophageal pathology in non-cardiac chest pain, and for colon cancer screening. As shown in Table 1, previous studies have focused on the use of deep learning for classifying gastrointestinal diseases and lesions identified on WCE images. Unsurprisingly, a frequently investigated outcome in published literature is bleeding. Deep learning models have enhanced WCE’s ability to detect bleeding lesions (including suspected blood content and angioectasia) with relatively high sensitivity and specificity[19-27]. In addition to bleeding, researchers have also used deep learning models in WCE to classify other gastrointestinal lesions such as ulcers[19-21,28-32], Crohn’s disease[33], polyps[7,19-21,34], celiac disease[6], and hookworm[35].
| Ref. | Class/outcome variable | Deep network architecture | Device/image resolution | Training and internal validation dataset | Testing/external validation dataset | Accuracy (%)/AUC | Sensitivity (%)/specificity (%) |
| Majid et al[19], 2020, NA | Multiple lesions (bleeding, esophagitis, ulcer, polyp) | CNN with classical features fusion and selection | NA/224 × 224 pixels | 70% of 12889 images from multiple databases | 30% of 12889 images from multiple databases | 96.5/NA | 96.5/NA |
| Ding et al[20], 2019, China | Multiple SB lesions1 | CNN (ResNet 152) | SB-CE by Ankon Technologies/480 × 480 pixels | 158235 images from 1970 patients | 113268334 images from 5000 patients | NA/NA | 99.88/100 (per patient); 99.90/100 (per lesion) |
| Iakovidis et al[21], 2018, NA | Multiple SB lesions2 | CNN and iterative cluster unification | (1) NA/489 × 409 pixels; and (2) MiroCam CE/320 × 320 pixels | (1) 465 images from 1063 volunteers; and (2) 852 images | (1) 233 images from 1063 volunteers; and (2) 344 images | (1) 89.9/0.963; and (2) 77.5/0.814 | (1) 90.7/88.2; and (2) 36.2/91.3 |
| Aoki et al[22], 2020, Japan | Bleeding (blood content) | CNN (ResNet50) | Pillcam SB2 or SB3 CE / 224 × 224 pixels | 27847 images from 41 patients | 10208 images from 25 patients | 99.89/0.9998 | 96.63/99.96 |
| Tsuboi et al[23], 2019, Japan | Bleeding (SB angioectasia) | CNN (SSD) | Pillcam SB2 or SB3 CE/300 × 300 pixels | 2237 images from 141 patients | 10488 images from 28 patients | NA/0.998 | 98.8/98.4 |
| Leenhardt et al[24], 2019, France | Bleeding (SB angioectasia) | CNN-based semantic segmentation | Pillcam SB3 CE / NA | 600 images | 600 images | NA/NA | 96/100 |
| Li et al[25], 2017, China | Bleeding (intestinal hemorrhage) | CNNs: (1) LeNet; (2) AlexNet; (3) GoogLeNet; and (4) VGG-Net | NA/NA | 9672 images | 2418 images | NA/NA | (1) 99.91/96.2; (2) 99.96/98.72; (3) 100/98.73; and (4) 99.96/98.72 |
| Jia et al[26], 2017, Hong Kong, China | Bleeding (both active and inactive) | CNN | NA/240 × 240 pixels | 1000 images | 500 images | NA/NA | 91.0/NA |
| Jia et al[27], 2016, Hong Kong, China | Bleeding (both active and inactive) | CNN (Inspired by AlexNet) | NA/240 × 240 pixels | 8200 images | 1800 images | NA/NA | 99.2/NA |
| Aoki et al[28], 2019, Japan | Ulcer (erosion or ulceration) | CNN (SSD) | Pillcam SB2 or SB3 CE/300 × 300 pixels | 5360 images from 115 patients | 10440 images from 65 patients | 90.8/0.958 | 88.2/90.9 |
| Wang et al[29], 2019, China | Ulcer | CNN (RetinaNet) | Magnetic-guided CE by Ankon Technologies/480 × 480 pixels | 37278 images from 1204 patient cases | 9924 images from 300 patient cases | 90.10/0.9469 | 89.71/90.48 |
| Wang et al[30], 2019, China | Ulcer | CNN (based on ResNet 34) | Magnetic-guided CE by Ankon Technologies/480 × 480 pixels | 80% of dataset from 1416 patients | 20% of dataset from 1416 patients | 92.05/0.9726 | 91.64/92.42 |
| Alaskar et al[31], 2019, NA | Ulcer | CNN: (1) GoogLeNet; and (2) AlexNet | NA /(1) 224 × 224 pixels; and (2) 227 × 227 pixels | 336 images | 105 images | (1) 100/1; and (2) 100/1 | (1) 100/100; and (2) 100/100 |
| Fan et al[32], 2018, China | (1) Ulcer; and (2) Erosion | CNN (AlexNet) | NA/511 × 511 pixels | (1) 5500 images; and (2) 7410 images | (1) 2750 images; and (2) 5500 images | (1) 95.16/0.9891; and (2) 95.34/0.9863 | (1) 96.80/94.79; and (2) 93.67/95.98 |
| Zhou et al[6], 2017, USA | Celiac disease | CNN (GoogLeNet) | Pillcam SB2 CE/512 × 512 pixels | 8800 images from 11 patients | 8000 images from 10 patients | NA/NA | 100/100 |
| Klang et al[33], 2020, Israel | Crohn’s disease | CNN (Xception) | Pillcam SB2 CE/299 × 299 pixels | Experiment 1: 80% of 17640 images from 49 patients; Experiment 2: Images from 48 patients | Experiment 1: 20% of 17,640 images from 49 patients; Experiment 2: Images from 1 individual patient | Experiment 1: 95.4-96.7/0.989-0.994; Experiment 2: 73.7–98.2/0.940-0.999 | Experiment 1: 92.5-97.1/96.0-98.1; Experiment 2: 69.5-100/56.8-100 |
| Saito et al[34], 2020, Japan | Polyp (protruding lesion) | CNN (SSD) | Pillcam SB2 or SB3 CE/300 × 300 pixels | 30584 images from 292 patients | 17507 images from 93 patients | 84.5/0.911 | 90.7/79.8 |
| Yuan et al[7], 2017, Hong Kong, China | Polyp | Deep neural network | Pillcam SB CE/64 × 64 pixels | Unknown proportion of 4000 images from 35 patients | Unknown proportion of 4000 images from 35 patients | 98/NA | 98/99 |
| He et al[35], 2018, Israel | Hookworm | CNN | Pillcam SB CE/227 × 227 pixels | 10 out of 11 patients (436796 images from 11 patients) | 1 individual patient (11-fold cross-validation) | 88.5/NA | 84.6/88.6 |
The deep network architecture is the full arrangement of neural networks in deep learning models covering input layer, hidden layers, and output layer. Although there were some variations with the deep network architecture, 16 out of 17 studies in Table 1 used CNN-based architectures in their deep learning models. The choice of deep network architectures depends on the classification objectives and individual research group. Nevertheless, many research groups prefer to use the well-known CNN-based architectures when classifying WCE images or benchmarking the performance of their custom deep learning architectures. These prebuilt CNN-based architects include LeNet[25], AlexNet[25,27,31,32], GoogLeNet[6,25,31], VGG-Net
In addition to variations in the deep learning architect, researchers had some variation in WCE device. There were three brands of WCE devices mentioned in these deep learning studies: PillCam (Medtronics), NaviCam (Ankon Technologies), and MiroCam (IntroMedic). Deep learning models can be incorporated with each device. However, different devices have different sizes and qualities of raw images, brightness, and camera angles. Since these devices are not standardized, the application of a specific deep learning model may not perform at the same prediction accuracy when applied universally to the other WCE devices.
Although the size and quality of the original WCE images is dependent on the device, image resolution is dependent on training time, deep network architecture, and lesion types. Intuitively, physicians prefer a higher image resolution when making an image-based diagnosis. However, higher image resolutions can lead to an increase in trainable parameters, floating-point operations, memory requirements, and training time. To counteract this, original images are often modified (either cropped or resized) to lower image resolution. As illustrated in Table 1, image resolution can range from 64 × 64 pixels to 512 × 512 pixels. The typical range of resolution is 240 × 240 pixels to 320 × 320 pixels. It is worth noting that all studies using the images captured by NaviCam (Ankon Technologies) selected the original image resolution of 480 × 480 pixels[20,29,30].
A collection of WCE images labeled by physicians is the main data source, which is commonly referred to as a dataset. As a part of data pre-processing, the dataset is typically divided into two groups. This creates two different datasets from the labeled WCE images. The first dataset is for training and internally validating the deep learning models. Once the final model is selected, the second dataset is used for testing the performance of the model with the data the model has not seen. Hence, data partitioning is one of the factors that could impact the predive performance of deep learning models[36].
There were two common approaches for dividing the initial dataset identified during the literature review. The first was to partition the data based on the aggregated images. The second was to partition the data per patient or video. The ratio of the two datasets varied dependent on the study, but common ratios included 50:50, 70:30 and 80:20[19,24,30,33]. The second approach to partition was often used when evaluating the predictive performance per patient[6,20,32,33]. Therefore, we can notice that the data partitioning approach in WCE images highly depends on the study design.
In medical literature, the most popular performance metrics are accuracy, sensitivity, specificity, and area under the curve (AUC). In the case of WCE images, where few WCE images are true lesions, accuracy and specificity can be skewed by deep learning models correctly identifying normal mucosa. For this reason, in data science, the focus on performance evaluation is on true positive classification[37]. In other words, data scientists prefer their models correctly classify the small number of positive images (e.g., angioectasia, tumor, or ulcer) rather than correctly classifying the normal mucosa images. Instead of accuracy and sensitivity, precision [true positive/(true positive + false positive)], recall [true positive/(true positive + false negative)], and F1 score (a harmonic mean of precision and recall) are the common performance metrics used by data scientists. It is worth noting that precision and recall are also known as positive predictive value and sensitivity respectively. Unfortunately, only a limited number of studies fully reported these set of performance metrics, especially F1 score[19,25-27]. In short, it is important to consider the performance metrics when determining or comparing the performance of deep learning models.
The main goal when analyzing WCE images is to detect abnormalities in the gastrointestinal tract. However, it is also helpful to detect normal mucosa and anatomical landmarks. As shown in Table 2, only two studies were designed to classify non-disease objects. The first study used deep learning to classify the complexities within the endoluminal scene, including turbid, bubbles, clear blob, wrinkles, and wall[38]. Although these images may not contribute to a final diagnosis, they can be used to characterize small intestine motility and to help rule out negative images. The second study created a predictive model for identifying organ locations such as the stomach, intestine, and colon[5]. Organ classification can be used to calculate the passage time of WCE in each organ and to determine if there are any motility disorders in the gastrointestinal tract. An important aspect of physician review of a WCE study is the identification of anatomical landmarks such as first images of the stomach, duodenum, and cecum which ultimately helps calculate capsule transit time through the small bowel. This transit time is vital to determining the location of lesion in the small bowel that may help guide treatment with deep enteroscopy techniques.
| Ref. | Class/outcome variable | Deep network architecture | Device/image resolution | Training and internal validation dataset | Testing/external validation dataset | Accuracy (%)/AUC | Sensitivity (%)/specificity (%) |
| Seguí et al[38], 2016, Spain | Scenes (turbid, bubbles, clear blob, wrinkles, wall) | CNN | Pillcam SB2 CE/100 × 100 pixels | 100000 images from 50 videos | 20000 images from 50 videos | 96/NA | NA/NA |
| Zou et al[5], 2015, NA | Organ locations (stomach, small intestine, and colon) | CNN (AlexNet) | NA/480 × 480 pixels | 60000 images | 15000 images | 95.52/NA | NA/NA |
An ideal goal for WCE would be the creation of a fully automated system for interpreting WCE images and generating accurate reports at least equivalent to conventional reading by physicians. Two retrospective studies compared the performance of conventional reading to the deep learning assisted reading (Table 3)[20,39]. The average reading times of deep learning assisted reading in both studies was less than 6 min. The average conventional reading time varied from 12 to 97 min depending on the expertise of the reader and the scope of WCE reading. In terms of overall lesion detection rate, there was a 3%-8% improvement of deep learning assisted reading over conventional reading. Interestingly, the accuracy of the deep learning model (as calculated during development) was higher than the actual detection rate. These findings may reflect the real-world challenges impacting human and deep learning model collaborations. An additional limitation was that there was no clear definition on how reading time was determined (e.g., from data preprocessing to final report generation).
| Ref. | Experiment type | Scope of WCE reading /device | Conventional reading | Deep learning assisted reading | P value |
| Aoki et al[39], 2019, Japan | Retrospective study using anonymized data | SB section only/Pillcam SB3 | mean reading time (min): Trainee: 20.7; Expert: 12.2 | mean reading time (min): Trainee: 5.2; Expert: 3.1 | < 0.001 |
| Overall lesion detection rate: Trainee: 47%; Expert: 84% | Overall lesion detection rate: Trainee: 55%; Expert: 87% | NS | |||
| Ding et al[20], 2019, China | Retrospective study by randomly selected videos | Small bowel abnormalities/SB-CE by Ankon Technologies | mean reading time ± standard deviation (min): 96.6 ± 22.53 | mean reading time ± standard deviation (min): 5.9 ± 2.23 | < 0.001 |
| Overall lesion detection rate: 41.43% | Overall lesion detection rate: 47.00% | NA1 |
The goal when creating a deep learning model is to best fit your target function. Overfitting is a classic problem that can occur after creating the initial deep learning model. Overfitting occurs when a model learns the detail and noise of the training data too well to the extent that it negatively impacts the performance of the model on new data. Despite the standard methods for dividing datasets during training and testing, the detection rate in deep learning assisted trials are not as good when compared to the rates during the initial training and testing process[20,39]. The decreased performance could indicate that the model fits the training dataset too closely and does not perform well with an unseen dataset. Another explanation could be imperfect human and machine collaboration. Since the human physician is the one who makes the final diagnosis based on the information provided by the deep learning model, the misdetection could be derived from how human physicians use or trust the judgment from deep learning models.
Traditionally, the risk stratification scores developed by one research team can be validated by another research team. Unfortunately, we have not seen the same level of transferability in deep learning research for WCE yet. As a result, the trials are very limited to their own research group and can be very difficult to have third party validation.
Each deep learning model is designed for a specific task that is based on the availability of positive lesions in their own dataset. Given this, it is questionable if it is even possible or effective to integrate these models. Integration can be even more complicated by the fact that each research group may use different devices, image resolutions, network architectures, and labeling practices.
One common barrier in medical device-related research is the use of proprietary file format. For example, the video file from PillCam device is stored in *.gvi and *.gvf file[40]. Thus, it may be difficult to extract data that is stored in the proprietary file format without help from the manufacturer. Such constraints may impact model integration and deployment. For example, it may take a longer time to prepare the files from deep learning models to use in a clinical setting. Also, there is no guarantee that the image resolution would be equivalent to the one seen in the proprietary reading software after extraction. For this reason, researchers should explore the pros and cons of each device available in their market to compare features and select the one that best aligns with their research goals.
Data preprocessing is the most time-consuming task in AI research. It is necessary to transform raw data into a ready-to-use and efficient format. Having a high-quality dataset is one of the key factors for creating a predictive model. By spending a lot of time extracting the data and labeling it, the dataset is a valuable asset to the research group. Ideally, high-quality datasets should be publicly available for researchers to use. However, there are a limited number of such datasets.
Since 2006, CNN-based architecture has proven to be an effective method for analyzing image data in various fields. Researchers have increasingly adopted CNN-based architecture for solving image classification problems. In our literature review, seventeen papers were identified that applied deep learning in WCE to classify gastrointestinal disorders. Our literature review demonstrated that the majority of CNN-based deep learning models were nearly perfect with regard to accuracy, sensitivity, specificity, and AUC[9].
There were only a few studies applying deep learning models to address non-disease objects, such as organ location and scenes in normal mucosa images (e.g., turbid, bubbles, clear blob, wrinkles, and wall). These non-disease objects are important building blocks toward a fully automated system and can aid in the identification of “landmarks” such as the first images of each bowel segment.
Although there seems to be an increasing amount of deep learning research on classifying WCE images, we are still in the early stages of investigating the utility of deep learning in enhancing clinical practice. The studies we identified often reflected the more standard view of WCE, as a means to view areas of the small bowel not accessible by upper and lower endoscopy. As the scope of WCE grows beyond the small bowel, we expect to see deep learning research on WCE expand accordingly. In addition, deep learning could enhance WCE capability to become highly effective in clinical practice and patient care by improving the speed and accuracy of WCE reading as well as predicting the location of abnormalities. Regardless of existing limitations and constraints, we expect the research and development in this area will continue to grow rapidly in the next decade.
The studies gathered in this literature review were indexed by PubMed. We also investigated publications concerning the utility of deep learning in computer science, medical image processing, mathematical modeling, and electrical engineering. Unfortunately, we cannot ensure that we identified every publication outside of PubMed.
In addition, it is difficult to compare one deep learning model to another based on their performance metrics alone. Most researchers have focused more on reporting traditional performance metrics without F1 score. The best practice for comparing these models would be to benchmark their performances on the same dataset that the models have never been trained on. To do so, researchers would need to make their trained models publicly available (e.g., uploading them to GitHub). This would allow clinical trials on deep learning models to expand outside their research group.
The idea of using computational algorithms for analyzing WCE images is not entirely new. The earliest study identified was published in 2006[41]. Universal to all these studies was a central hypothesis investigating the ability of computational algorithms to improve the efficiency of reading WCE studies, specifically in terms of time and accuracy. The prospect of a fully automated system for interpreting WCE images would benefit patient care because of fast and accurate diagnoses of gastrointestinal medical conditions such as bleeding, polyps, Crohn’s disease, and cancer.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kawabata H S-Editor: Wang JL L-Editor: A P-Editor: Wang LL
| 1. | Moglia A, Menciassi A, Schurr MO, Dario P. Wireless capsule endoscopy: from diagnostic devices to multipurpose robotic systems. Biomed Microdevices. 2007;9:235-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 89] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 2. | Li B, Meng MQ, Xu L. A comparative study of shape features for polyp detection in wireless capsule endoscopy images. Annu Int Conf IEEE Eng Med Biol Soc. 2009;2009:3731-3734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 3. | Mishkin DS, Chuttani R, Croffie J, Disario J, Liu J, Shah R, Somogyi L, Tierney W, Song LM, Petersen BT; Technology Assessment Committee; American Society for Gastrointestinal Endoscopy. ASGE Technology Status Evaluation Report: wireless capsule endoscopy. Gastrointest Endosc. 2006;63:539-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 169] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 4. | Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, Ohnishi T, Fujishiro M, Matsuo K, Fujisaki J, Tada T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 426] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 5. | Zou Y, Li L, Wang Y, Yu J, Li Y, Deng W. Classifying digestive organs in wireless capsule endoscopy images based on deep convolutional neural network. Proceedings of the 2015 IEEE International Conference on Digital Signal Processing (DSP); 2015 July 21-24; Singapore, Singapore. IEEE, 2015: 1274-1278. [DOI] [Full Text] |
| 6. | Zhou T, Han G, Li BN, Lin Z, Ciaccio EJ, Green PH, Qin J. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput Biol Med. 2017;85:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 7. | Yuan Y, Meng MQ. Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys. 2017;44:1379-1389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 8. | Sahiner B, Pezeshk A, Hadjiiski LM, Wang X, Drukker K, Cha KH, Summers RM, Giger ML. Deep learning in medical imaging and radiation therapy. Med Phys. 2019;46:e1-e36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 516] [Cited by in RCA: 402] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 9. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology 2020; 158: 76-94. e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 323] [Article Influence: 64.6] [Reference Citation Analysis (1)] |
| 10. | Ruffle JK, Farmer AD, Aziz Q. Artificial Intelligence-Assisted Gastroenterology- Promises and Pitfalls. Am J Gastroenterol. 2019;114:422-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 11. | Thakkar SJ, Kochhar GS. Artificial intelligence for real-time detection of early esophageal cancer: another set of eyes to better visualize. Gastrointest Endosc. 2020;91:52-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 12. | Mahmud M, Kaiser MS, Hussain A, Vassanelli S. Applications of Deep Learning and Reinforcement Learning to Biological Data. IEEE Trans Neural Netw Learn Syst. 2018;29:2063-2079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 245] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 13. | Spann A, Yasodhara A, Kang J, Watt K, Wang B, Goldenberg A, Bhat M. Applying Machine Learning in Liver Disease and Transplantation: A Comprehensive Review. Hepatology. 2020;71:1093-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 111] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 14. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology 2018; 155: 1069-1078. e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 432] [Article Influence: 61.7] [Reference Citation Analysis (1)] |
| 15. | Kang R, Park B, Eady M, Ouyang Q, Chen K. Single-cell classification of foodborne pathogens using hyperspectral microscope imaging coupled with deep learning frameworks. Sens Actuators B Chem. 2020;309:127789. [DOI] [Full Text] |
| 16. | Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O. Deep learning with convolutional neural network in radiology. Jpn J Radiol. 2018;36:257-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 216] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 17. | Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak JAWM; the CAMELYON16 Consortium, Hermsen M, Manson QF, Balkenhol M, Geessink O, Stathonikos N, van Dijk MC, Bult P, Beca F, Beck AH, Wang D, Khosla A, Gargeya R, Irshad H, Zhong A, Dou Q, Li Q, Chen H, Lin HJ, Heng PA, Haß C, Bruni E, Wong Q, Halici U, Öner MÜ, Cetin-Atalay R, Berseth M, Khvatkov V, Vylegzhanin A, Kraus O, Shaban M, Rajpoot N, Awan R, Sirinukunwattana K, Qaiser T, Tsang YW, Tellez D, Annuscheit J, Hufnagl P, Valkonen M, Kartasalo K, Latonen L, Ruusuvuori P, Liimatainen K, Albarqouni S, Mungal B, George A, Demirci S, Navab N, Watanabe S, Seno S, Takenaka Y, Matsuda H, Ahmady Phoulady H, Kovalev V, Kalinovsky A, Liauchuk V, Bueno G, Fernandez-Carrobles MM, Serrano I, Deniz O, Racoceanu D, Venâncio R. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA. 2017;318:2199-2210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1871] [Cited by in RCA: 1556] [Article Influence: 194.5] [Reference Citation Analysis (0)] |
| 18. | Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5683] [Cited by in RCA: 5353] [Article Influence: 669.1] [Reference Citation Analysis (0)] |
| 19. | Majid A, Khan MA, Yasmin M, Rehman A, Yousafzai A, Tariq U. Classification of stomach infections: A paradigm of convolutional neural network along with classical features fusion and selection. Microsc Res Tech. 2020;83:562-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 82] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 20. | Ding Z, Shi H, Zhang H, Meng L, Fan M, Han C, Zhang K, Ming F, Xie X, Liu H, Liu J, Lin R, Hou X. Gastroenterologist-Level Identification of Small-Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-Learning Model. Gastroenterology 2019; 157: 1044-1054. e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 208] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 21. | Iakovidis DK, Georgakopoulos SV, Vasilakakis M, Koulaouzidis A, Plagianakos VP. Detecting and Locating Gastrointestinal Anomalies Using Deep Learning and Iterative Cluster Unification. IEEE Trans Med Imaging. 2018;37:2196-2210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 22. | Aoki T, Yamada A, Kato Y, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Automatic detection of blood content in capsule endoscopy images based on a deep convolutional neural network. J Gastroenterol Hepatol. 2020;35:1196-1200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 23. | Tsuboi A, Oka S, Aoyama K, Saito H, Aoki T, Yamada A, Matsuda T, Fujishiro M, Ishihara S, Nakahori M, Koike K, Tanaka S, Tada T. Artificial intelligence using a convolutional neural network for automatic detection of small-bowel angioectasia in capsule endoscopy images. Dig Endosc. 2020;32:382-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 24. | Leenhardt R, Vasseur P, Li C, Saurin JC, Rahmi G, Cholet F, Becq A, Marteau P, Histace A, Dray X; CAD-CAP Database Working Group. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest Endosc. 2019;89:189-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (1)] |
| 25. | Li P, Li Z, Gao F, Wan L, Yu J. Convolutional neural networks for intestinal hemorrhage detection in wireless capsule endoscopy images. Proceedings of the 2017 IEEE International Conference on Multimedia and Expo (ICME); 2017 July 10-14; Hong Kong, China. IEEE, 2017: 1518-1523. [DOI] [Full Text] |
| 26. | Jia X, Meng MQ. Gastrointestinal bleeding detection in wireless capsule endoscopy images using handcrafted and CNN features. Annu Int Conf IEEE Eng Med Biol Soc. 2017;2017:3154-3157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 27. | Jia X, Meng MQ. A deep convolutional neural network for bleeding detection in Wireless Capsule Endoscopy images. Annu Int Conf IEEE Eng Med Biol Soc. 2016;2016:639-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 28. | Aoki T, Yamada A, Aoyama K, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Tanaka S, Koike K, Tada T. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc 2019; 89: 357-363. e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 174] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 29. | Wang S, Xing Y, Zhang L, Gao H, Zhang H. A systematic evaluation and optimization of automatic detection of ulcers in wireless capsule endoscopy on a large dataset using deep convolutional neural networks. Phys Med Biol. 2019;64:235014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 30. | Wang S, Xing Y, Zhang L, Gao H, Zhang H. Deep Convolutional Neural Network for Ulcer Recognition in Wireless Capsule Endoscopy: Experimental Feasibility and Optimization. Comput Math Methods Med. 2019;2019:7546215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 31. | Alaskar H, Hussain A, Al-Aseem N, Liatsis P, Al-Jumeily D. Application of Convolutional Neural Networks for Automated Ulcer Detection in Wireless Capsule Endoscopy Images. Sensors (Basel). 2019;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 32. | Fan S, Xu L, Fan Y, Wei K, Li L. Computer-aided detection of small intestinal ulcer and erosion in wireless capsule endoscopy images. Phys Med Biol. 2018;63:165001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 33. | Klang E, Barash Y, Margalit RY, Soffer S, Shimon O, Albshesh A, Ben-Horin S, Amitai MM, Eliakim R, Kopylov U. Deep learning algorithms for automated detection of Crohn's disease ulcers by video capsule endoscopy. Gastrointest Endosc 2020; 91: 606-613. e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 34. | Saito H, Aoki T, Aoyama K, Kato Y, Tsuboi A, Yamada A, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc 2020; 92: 144-151. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 109] [Article Influence: 21.8] [Reference Citation Analysis (1)] |
| 35. | He JY, Wu X, Jiang YG, Peng Q, Jain R. Hookworm Detection in Wireless Capsule Endoscopy Images With Deep Learning. IEEE Trans Image Process. 2018;27:2379-2392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 36. | Korjus K, Hebart MN, Vicente R. An Efficient Data Partitioning to Improve Classification Performance While Keeping Parameters Interpretable. PLoS One. 2016;11:e0161788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 37. | Sokolova M, Lapalme G. A systematic analysis of performance measures for classification tasks. Inf Process Manag. 2009;45:427-437. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2621] [Cited by in RCA: 1507] [Article Influence: 94.2] [Reference Citation Analysis (0)] |
| 38. | Seguí S, Drozdzal M, Pascual G, Radeva P, Malagelada C, Azpiroz F, Vitrià J. Generic feature learning for wireless capsule endoscopy analysis. Comput Biol Med. 2016;79:163-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 39. | Aoki T, Yamada A, Aoyama K, Saito H, Fujisawa G, Odawara N, Kondo R, Tsuboi A, Ishibashi R, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Clinical usefulness of a deep learning-based system as the first screening on small-bowel capsule endoscopy reading. Dig Endosc. 2020;32:585-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 40. | PillCam® Capsule Endoscopy. User Manual RAPID® v9.0, DOC-2928-02, Novermber 2016, p.123. Available from: https://www.pillcamcrohnscapsule.eu/assets/pdf/DOC-2928-02-PillCam-Desktop-SWv9UMEN.pdf. |
| 41. | Spyridonos P, Vilariño F, Vitrià J, Azpiroz F, Radeva P. Anisotropic feature extraction from endoluminal images for detection of intestinal contractions. Med Image Comput Comput Assist Interv. 2006;9:161-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |