Published online Jul 28, 2020. doi: 10.37126/aige.v1.i1.6
Peer-review started: June 23, 2020
First decision: July 3, 2020
Revised: July 7, 2020
Accepted: July 17, 2020
Article in press: July 17, 2020
Published online: July 28, 2020
Processing time: 30 Days and 3.3 Hours
Artificial intelligence (AI) allows machines to provide disruptive value in several industries and applications. Applications of AI techniques, specifically machine learning and more recently deep learning, are arising in gastroenterology. Computer-aided diagnosis for upper gastrointestinal endoscopy has growing attention for automated and accurate identification of dysplasia in Barrett’s esophagus, as well as for the detection of early gastric cancers (GCs), therefore preventing esophageal and gastric malignancies. Besides, convoluted neural network technology can accurately assess Helicobacter pylori (H. pylori) infection during standard endoscopy without the need for biopsies, thus, reducing gastric cancer risk. AI can potentially be applied during colonoscopy to automatically discover colorectal polyps and differentiate between neoplastic and non-neoplastic ones, with the possible ability to improve adenoma detection rate, which changes broadly among endoscopists performing screening colonoscopies. In addition, AI permits to establish the feasibility of curative endoscopic resection of large colonic lesions based on the pit pattern characteristics. The aim of this review is to analyze current evidence from the literature, supporting recent technologies of AI both in upper and lower gastrointestinal diseases, including Barrett's esophagus, GC, H. pylori infection, colonic polyps and colon cancer.
Core tip: Artificial intelligence (AI) allows machines to provide disruptive value in a multitude of industries and knowledge domains. Applications of artificial intelligence techniques, specifically machine learning and more recently deep learning, are arising in gastrointestinal endoscopy. Computer-aided diagnosis has been performed during upper gastrointestinal endoscopy for the automated identification of dysplastic lesions in Barrett’s esophagus for preventing esophageal cancer, as well as in lower gastrointestinal endoscopy for detecting colorectal polyps to prevent colorectal cancer. The aim of this review is to investigate current data from the literature, supporting recent technologies of AI both in upper and lower gastrointestinal diseases, including Barrett's esophagus, gastric cancer, Helicobacter pylori infection, colonic polyps and colon cancer.
- Citation: Morreale GC, Sinagra E, Vitello A, Shahini E, Shahini E, Maida M. Emerging artificial intelligence applications in gastroenterology: A review of the literature. Artif Intell Gastrointest Endosc 2020; 1(1): 6-18
- URL: https://www.wjgnet.com/2689-7164/full/v1/i1/6.htm
- DOI: https://dx.doi.org/10.37126/aige.v1.i1.6
Artificial intelligence (AI) is based on intelligent agents performing functions associated with human mind, such as learning and problem solving[1,2].
In endoscopy, AI has begun to assist the improvement of colonic polyp detection and adenoma detection rate (ADR), to discriminate between benign and precancerous lesions based on the interpretation of their superficial patterns.
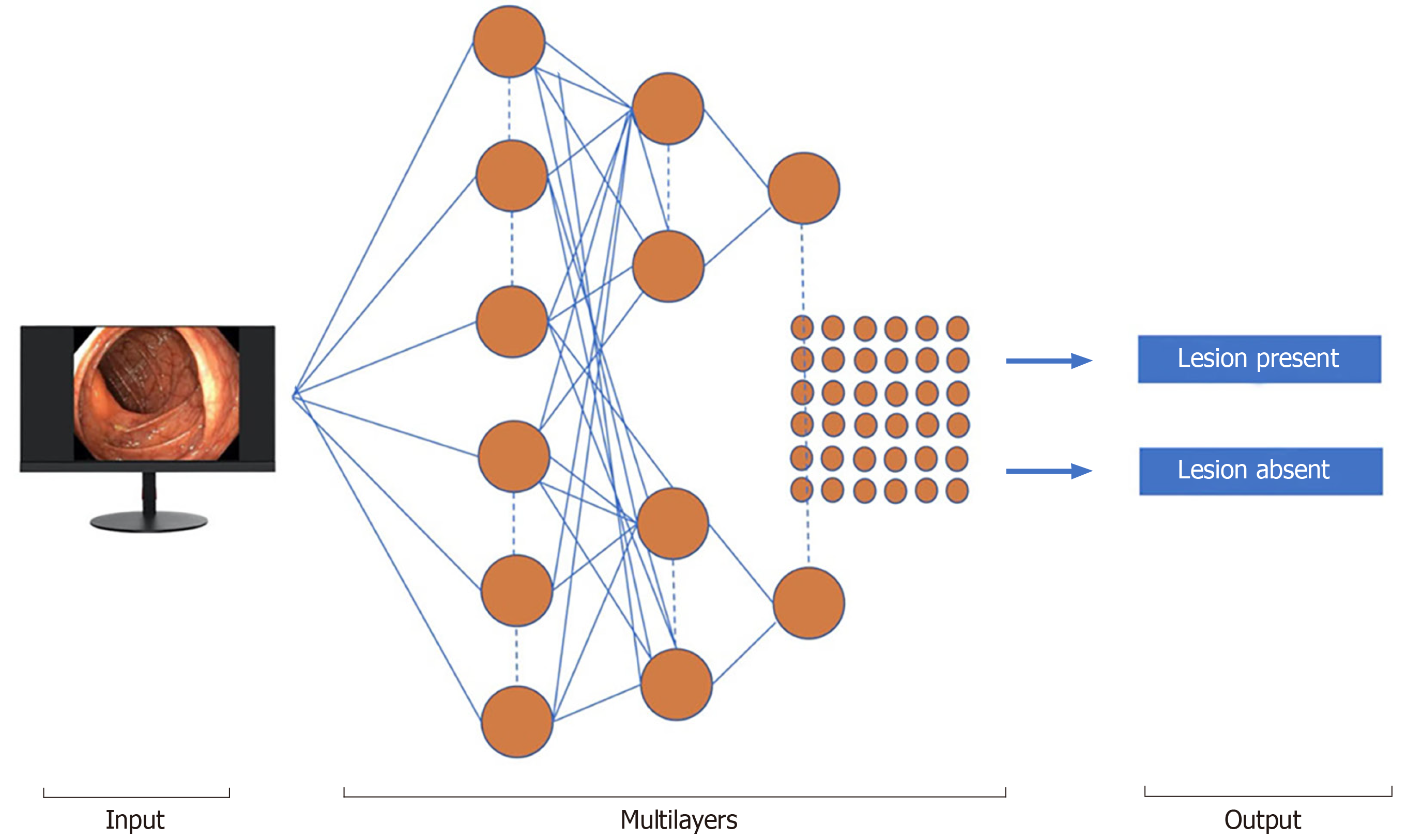
Machine learning (ML) and deep learning (DL) can be considered subfields of AI. ML is a form of AI that can support decision process allowing the improvement, without any Programmation, of the algorithms applied, including data testing and the implementation of descriptive and predictive models (Figure 1).
ML is distinguished into supervised and unsupervised methods. An instance of supervised ML, artificial neural networks (ANN), mirror the scheme function of the brain. Each neuron is a computing unit and all neurons are connected to produce a network. ML and convoluted neural network (CNN) algorithms have been created to train software to discriminate normal from abnormal regions in the lumen of the gut. For polyp detection, ML uses a fixed number of characteristics, such as polyp size, shape, and mucosal patterns.
A variety of deep learning neural network architectures are included in DL-based methods that automatically extract relevant imaging features without the human perceptual biases[3].
Barrett's esophagus (BE) is characterized by an unusual (metaplastic) transformation of the mucosal cells, lining the lower part of the esophagus, from normal stratified squamous epithelium to columnar one and associated with interspersed goblet cells[4]. This condition represents a risk factor for esophageal adenocarcinoma (EAC) whose most serious prognosis is related to the late diagnosis[4]. Moreover, 93% of patients can achieve a complete disease remission after a regular surveillance during 10 years and treatment[5-7]. Promising techniques for the management of BE with the potential of reducing the cancer risk by an accurate diagnosis of dysplasia, are being developed.
However, despite some limitations in interventional therapies, such as endoscopic resection (ER) and ablation techniques (radiofrequency ablation or cryoablation) they can help preventing the evolution into malignancy[8-11].
The recognition of neoplastic changes in BE patients is crucial and innovations in endoscopic imaging have worked for early detection of minimal epithelial neoplastic lesions based on distinct mucosal features.
In a first study, Mendel et al[12], introduced a useful method for generating an automatic classification based on endoscopic white light images through the learning of specific features helped by a pretrained deep residual network, instead of handcrafted texture features. The study used a data set of 100 high-resolution endoscopic images from 39 patients supplied by the Endoscopic Vision Challenge Medical Image Computing and Computer-Assisted Intervention (MICCAI). While 22 BE patients had cancerous lesions, 17 had non-cancerous BE.
The endoscopic images were independently evaluated by five experts and then compared with probability maps provided by AI, showing a strong correspondence. Since the significant of manual segmentations vary significantly, their intersection was considered as a cancerous region (C1-region) within each C1-image.
Ebigbo et al[13], employed two data sets to train and validate a computer-aided diagnosis (CAD) system relying on a deep CNN with a residual net (ResNet) architecture. Images consisted of 148 high-definition white light endoscopy (WLE) and narrowband imaging (NBI) images regarding 33 EAC and 41 areas of non-neoplastic BE in the Augsburg data set, while the MICCAI data set comprised 100 high-definition WLE images, 17 early EAC and 22 areas of non-neoplastic BE. CAD-DL system diagnosed EAC with a sensitivity of 97% and a specificity of 88% for WLE images, whereas a sensitivity and specificity of 94% and 80% for NBI images, respectively. CAD-DL reached a sensitivity and specificity of 92% and 100%, respectively, for the MICCAI images.
In these beginning studies, the authors developed a CAD model and displayed promising performance scores in the classification/segmentation areas during BE assessment.
However, these results were achieved using high-quality endoscopic imaging that cannot always be obtained during daily clinical practice. This system was previously developed to further increase the speed of image analysis for classification and the resolution of the dense prediction, displaying the color-coded spatial distribution of cancer probabilities.
Still based on deep CNNs and a ResNet architecture with DeepLab V.3+, a state-of-the-art encoder-decoder network was readjusted. To transfer the endoscopic Livestream to our AI system, a capture card (Avermedia, Taiwan) for image aquisition was incorporated into the endoscopic monitor[14] and the AI system was trained by using 129 endoscopic images. All AI-image outcomes were confirmed by pathological examination of resection specimens (EAC), as well as forceps biopsies (i.e., normal BE). The AI system showed high performance scores in the categorization task with a sensitivity and specificity of 83.7% and 100%, respectively.
CNN was also used by Horie et al[15], that retrospectively collected 8428 training images from esophageal cancer of 384 patients through CNNs. CNN took 27 seconds to analyze 1118 test images and correctly detected esophageal cancer cases with a sensitivity of 98%. CNN detected every 7 small cancer lesions lower than 10 mm in size. This system facilitated early and rapid malignancy detection leading to a better prognosis of these patients.
AI can assist endoscopists to make targeted biopsies with high-accuracy, saving work/time-intensive random sampling, with a low sensitivity (64%) for the detection of dysplasia. An international, randomized, crossover trial[16], compared high-definition white-light endoscopy (HD-WLE) and NBI for detecting IM and malignancy in 123 patients with BE (mean circumferential and maximal sizes, 1.8 and 3.6 cm, respectively).
Both HD-WLE and NBI detected 104/113 (92%) patients with IM, but NBI required fewer biopsies per-patient and exhibited a significantly higher dysplasia detection rate (30% vs 21%). During endoscopic examination with NBI, all areas of HGD and cancer presented an irregular mucosal or vascular pattern. Regular NBI surface patterns did not harbor HGD or cancer, suggesting that biopsies could be potentially avoided in the latter cases. Besides, in a multicenter, randomized crossover study[17], using endoscopic trimodal imaging (ETMI) for detection of early neoplasia in BE, ETMI showed no improvement in overall dysplasia detection than standard video endoscopy. The diagnosis of dysplasia was still made in a significant number of patients by random biopsies, and patients with a confirmed diagnosis of LGIN had a significant risk of HGIN/carcinoma.
Van der Sommen et al[18] used a computer algorithm to detect early neoplastic lesions in BE and employed specific texture, color filters, and ML-based on 100 images from 44 patients with BE. This system identified early neoplastic lesions on a patient-level with a sensitivity and specificity of 86% and 87%, respectively. The author assumed that the automated computer algorithm implemented for this study was able to identify early neoplastic lesions with reasonable accuracy.
De Groof et al[19] developed a CAD system using endoscopic images of Barrett's neoplasm based on the endoscopic images of 40 Barrett's neoplastic lesions and 20 non-dysplastic BE, reaching a sensitivity and specificity for the detection of such lesions of 95% and 85%, respectively.
AI technology was applied for volumetric laser endomicroscopy (VLE) in 2017. VLE with laser marking is a broad field of advanced imaging technology that was commercially available in the United States in 2013 to facilitate dysplasia detection.
VLE can enhance the detection of neoplastic lesions in BE by performing a circumferential scan of the esophageal wall layers. Sixteen patients with BE were included in the study and a total of 222 laser markers (LMs) were placed, 97% of them were visible on WLE. All LMs were evident on VLE directly after marking, and 86% were confirmed during the post hoc analysis. LM targeting held an accuracy of 85% of cautery marks. This original study applied to humans showed that VLE-guided LM can be a possible and secure procedure[20].
In another study[21] the same authors used a database of VLE images from BE endoscopic resection specimens with/without neoplasia, precisely correlated them with histology to develop a VLE prediction score. The receiving operating characteristic curve of this prediction score showed an area under the curve (AUC) of 0.81. A value ≥ 8 correlated with an 83% sensitivity and 71% specificity.
Optical coherence tomography (OCT) is a technique that produces high-resolution esophageal images through endoscopy. OCT can recognize specialized IM from epithelial squamous cells, but image criteria for distinguishing intramucosal carcinoma (IMC) and HGD from LGD, indeterminate-grade dysplasia (IGD), and specialized IM without dysplasia have not been approved yet.
Evans et al[22], examined 177 OCT images from patients with a histological diagnosis of BE. The histopathology analysis was IMC/HGD in 49 cases, LGD in 15, IGD in 8, specialized IM in 100, whereas gastric mucosa in 5 patients. A meaningful correlation was found between the MC/HGD histopathologic result and scores for each image feature, surface maturation, and gland architecture. When a dysplasia index determination of ≥ 2 was used, an 83% sensitivity and 75% specificity were determined for diagnosing IMC/HGD.
In a tertiary-care center, 27 BE patients underwent 50 EMRs imaged by VLE and pCLE, and were classified into neoplastic/non-neoplastic on the basis of histology result. The sensitivity and specificity of pCLE for detecting BE dysplasia, was 76% and 79%, respectively. The OCT-SI showed a sensitivity of 70% and a specificity of 60%. Moreover, the novel VLE-DA showed a sensitivity of 86%, specificity of 88% and a diagnostic accuracy of 87%[23].
Esophageal squamous cell carcinoma (SCC) is the sixth malignant cause of mortality worldwide and a greater percentage affect developing countries due to a delayed diagnosis[24]. Lugol's chromoendoscopy currently represents the gold standard technique for identifying SCC during gastroscopy, despite a low specificity (about 70%) but a higher sensitivity (over 90%).
Among non-invasive tests, NBI is another approach that has a low diagnostic specificity as displayed in a randomized controlled trial (RCT), related to the physician’s experience[25].
High-resolution microendoscopy (HRME) has shown the potential to enhance esophageal SCC detection during screening. An automated, real-time analysis algorithm has been developed and assessed using training tests, and validation images derived from a previous in-vivo study including 177 subjects involved for screening/surveillance programs. In a post hoc analysis, the algorithm recognized malignant tumors with a 95% sensitivity and 91% specificity, in the validation dataset, while 84% and 95% in the original study. Therefore, this technology could be applied in settings with less expertise operators in interpreting HRME images[26].
Kodashima et al[27] realized a computer system architecture to simplify the differentiation among neoplastic features and healthy tissues as a result of analyzing images in endocytoscopy of esophageal tissue from histopathological analysis, by analyzing the nuclear area of the collected images from 10 patients, to achieve an accurate and automatic diagnosis[27].
Shin et al[28] developed a quantitative image analysis algorithm that was able to recognize squamous dysplasia from non-neoplastic mucosa. They completed an image interpretation of 177 subjects undergoing upper endoscopy for SCC screening or surveillance, by using HRME. Quantitative data from the high-resolution images were used to create an algorithm to identify high-grade squamous dysplastic lesions or invasive SCC on histopathology.
The highest performance was gained using the mean nuclear area as the input for classification, resulting in a sensitivity and specificity of 93% and 92% in the training set, 87% and 97% in the test set, 84% and 95% in an independent validation set, respectively. ER is a technique employed for treating tumors with submucosal invasion depth 1 (SM1), whereas surgical removal with/without chemo-radiotherapy is usually used for SCC cases with a tumor infiltration deeper than SM2.
Accordingly, the preoperative endoscopic estimation of the ESCC invasion depth is critical. Recently, a rapid improvement in the application of AI with DL in medicine has been realized. A study by Tokai et al[29], evaluated the efficacy of AI in measuring ESCC invasion depth in a set of 1751 ESCC training images. AI recognized 95.5% (279/291) of the ESCC in the 10 test images when analyzing the 279 images it correctly predicted the invasion depth of the ESCC with an 84.1% sensitivity and an 80.9% accuracy in 6 seconds, much more precise for the estimation of ESCC invasion depth from endoscopists.
Gastric cancer (GC) ranks third main cause of malignancy mortality worldwide, and esophagogastroduodenoscopy (EGD) is considered the best diagnostic tool for neoplasms at their early stages. The treatment of gastric tumors depends on the depth of the submucosal invasion; indeed, for differentiated intramucosal tumors (M) or those that invade the superficial submucosal layer (≤ 500 lm: SM1) ER is provided, while those with a deep submucosal invasion (> 500 lm: SM2) should be surgically treated for the potential risk of local invasiveness and metastases. Magnifying endoscopy combined with NBI or FICE (flexible color enhancement of spectral imaging) is clinically useful in discriminating gastric malignant from non-malignant areas[30-34]. However, this optical diagnosis strictly depends on the expertise and the experience of the operator, which prevents its general use in clinical practice.
Two RCTs examined the performance of endoscopy with/without the support of AI algorithms. The first research estimated the performance of a real-time DL system, WISENSE, to control the presence of blind spots during EGD. Overall, 324 patients randomly performed endoscopy with or without the use of WISENSE that monitored blind spots with a 90% average accuracy, and a separate accuracy for each site ranging 70.2%-100% in the 107 live endoscopic videos.
The average sensitivity and specificity were 87.6% and 95%, ranging between 63.4%-100% and 75%-100%, respectively. For timing endoscopic procedure, WISENSE accurately predicted the start and end times in 93.5% (100/107) and 97.2% (104/107) videos, respectively[35].
Miyaki et al[36], developed software allowing a quantitative evaluation of mucosal GCs on magnifying gastrointestinal endoscopy images obtained with FICE. They adopted a set of features framework having densely sampled scale-invariant feature transform descriptors to magnifying FICE images of 46 intramucosal GCs then compared with histologic findings. The CAD system allowed an 86%detection accuracy, a sensitivity and specificity of 85% and 87% for a cancer diagnosis, respectively.
In the study by Kanesaka et al[37], a total of 127 patients with EGC contributed to 127 cancerous M-NBI images, while 20 not-EGC patients provided to 60 not-cancerous M-NBI images. The authors created software that allowed both the identification of GC and outlined the edge between malignant and non-malignant regions. This CAD algorithm was designed to investigate grey-level co-occurrence matrix characteristics of partitioned pixel slices of magnifying NBI images, and a support vector machine was used for the ML method. The models showed a 97% sensitivity and 95% specificity in distinguishing cancer, while the performance for area concordance displayed a sensitivity and specificity, of 81% and 66% respectively.
In 2018, Hirasawa et al[38], elaborated an AI-based diagnostic system to detect GC, using a CNN simulating the human brain.
A total of 714 among 2,296 test image sets (31.1%) confirmed GC presence, and 84.1% had moderate/severe gastric atrophy. The CNN employed 47 seconds to analyze the 2,296 test images, diagnosing overall 232 GCs, 161 as non-malignant lesions, 71 of 77 as GC lesions with a sensitivity of 92.2%. The majority of gastric lesions (98.6%) with a diameter ≥ 6 mm were precisely identified by CNN, additionally to all invasive carcinomas (T1b or deeper). The undiagnosed lesions had a superficial depression and were more frequently intramucosal cancers with a differentiated-histotype, whose discrimination from gastric inflammation was challenging also for experienced endoscopists. Another usual reason for misdiagnosis was the anatomical sites of the cardia, incisura angularis, and pylorus.
Zhu et al[39] examined the potential of AI to address the prediction of invasion depth of early GC. In particular, they developed and validated an AI model CNN-CAD that used a deep learning algorithm for determining EGC invasion depth (“M/SM1” vs “SM2 or deeper”).
A total of 790 endoscopic images of GCs were employed for ML, while an additional 203 images, completely autonomous from the learning material, were handled as a test set. The AI model exhibited a sensitivity and specificity of 76% and 96%, respectively in distinguishing SM2 or deeper cancer invasion, with a higher diagnostic performance as compared to the one reached by endoscopists. This high specificity could lessen the overestimation of tumoral invasion, which would contribute indirectly to reduce avoidable surgeries for M/SM1 malignancies. Moreover, in this study, the CNN-CAD system also achieved significantly greater accuracy and specificity than both expert and junior trained endoscopists.
AI might assist physicians to predict prognoses of patients with GC. Some crucial clinical trials evaluating adjuvant strategies of advanced GC were produced over the past decade, but the most suitable therapy for GC is so far uncertain. Besides, two contemporary molecular landscape studies proved the presence of various molecular GC subtypes[40,41].
A DL-based model (survival recurrent network, SRN) was developed to predict survival events for a total of 1190 GC patients, based on clinical/pathology data as well as therapy regimens, predicting the outcome at each-time point during a 5-year surveillance time.
The SRN showed that the mesenchymal subtype of GC should stimulate a tailored postoperative therapeutical strategy as a consequence of its great risk of recurrence rate. Conversely, the SRN observed that GCs with microsatellite instability and the papillary type displayed significantly more favorable prognosis after chemotherapy including capecitabine and cisplatin. SRN reached a survival of 92%, 5 years after curative gastrectomy resection[42].
ANN model was used to evaluate 452 GC patients, determining survival times with approximately 90% accuracy, and focusing on producing an adequate ANN structure with the capacity to handle censored data[43]. In detail, 5 sets of single time-point feed-forward ANN models were generated to predict the outcomes of GC patients at regular time intervals (every year) until the fifth year after gastrectomy. Hence, the ANN prediction models exhibited accuracy, sensitivity, and specificity ranging as follows 88.7%-90.2%, 70.2%-92.5%, and 66.7%-96.2%, respectively.
Helicobacter pylori (H. pylori) infects the epithelial gastric cells and is associated with functional dyspepsia, peptic ulcers, mucosal atrophy, intestinal metaplasia, and GC[44]. H. pylori-associated chronic gastritis may also raise the risk of GC[45,46]. CNN technology can accurately assess H. pylori infection during conventional endoscopy without needing biopsies. In a pilot study by Zheng et al[47], the authors produced a Computer-Aided Decision Support System that uses CNN to estimate H. pylori infection based on endoscopic images. From 1959 patients, 77% were assigned to the derivation cohort (1507 patients; 11729 gastric images) and 56% of them had H. pylori infection (847), while 23% were selected for the validation cohort (452) and 69% of patients were H. pylori infected (310; 3755 total images).
Huang et al[48] applied neural networks (refined feature selection with a neural network, RFSNN) to predict H. pylori-related gastric histological hallmarks based on standard endoscopic images. The authors trained the model using endoscopic images of 30 patients and used image parameters taken from a different cohort of 74 patients to generate a model to predict H. pylori infection, showing an 85% sensitivity and a 91% specificity for identifying H. pylori infection. Moreover, RFSNN revealed an accuracy higher than 80% in predicting the presence of gastric atrophy, IM, and H. pylori-related gastritis severity.
Shichijo et al[49] produced a 22-layer deep CNN to predict H. pylori infection during real-time endoscopy. A dataset including 32208 images of 735 H. pylori-positive and 1015 H. pylori-negative patients was handled. The sensitivity/specificity/accuracy, were 81.9/83.4/83.1%, respectively, for the first CNN, and 88.9/87.4/87.7%, respectively, for the secondary CNN, employing in both cases a similar time (198 seconds and 194 seconds, respectively).
Another study group developed a CNN, preparing 179 endoscopic images obtained from 139 patients (65 were H. pylori-positive and 74 H. pylori-negative). One hundred and fifty-nine of all images were adopted as training for a standard neural network, and the remaining 30 (15 of H. pylori-negative and 15 of H. pylori-positive patients) as test images. CAD model showed an 87% sensitivity and specificity to detect H. pylori infection with an AUC of 0.96[50].
Nakashima et al[51] used blue laser images (BLI)-bright and linked color imaging (LCI) on 162 patients as learning material and those from 60 patients as a test data set. From each patient, three white-light images (WLI), three BLI, and three linked color images (LCI; Fujifilm Corp.) were obtained, respectively. For WLI, the AUC was 0.66.
Colorectal cancer (CRC) is the third most frequent malignancy in males and second in females, and the fourth most frequent cause of cancer fatality[52]. The National Polyp Study registered that 70%-90% of CRCs can be prevented by routine endoscopic surveillance and removal of polyps[53], but 7%-9% of CRCs can occur despite these measures[54].
Around 85% of “interval cancers” are due to missed polyps or inadequately removed polyps[55]. Adenomas are the most common precancerous lesions throughout the colon. The ADR measures the endoscopist ability to identify adenomas. The ADR ranges between 7%–53% among endoscopists making depending on their training, endoscopic removal technique, withdrawal time, quality of bowel preparation, and other procedure-dependent determinants[56,57].
Several endoscopic innovations have been promoted to increase the ADR[58,59].
A review including 5 studies on the effect of high-resolution colonoscopes on the ADR showed conflicting results; a study concluded that the ADR is raised exclusively for endoscopists with an ADR lower than 20%[60].
CAD analysis has the potential to aid adenoma detection further.
Urban et al[61], used a different and representative set of 8641 hand-labeled images from screening colonoscopies handled among over 2000 patients. They tested the models on 20 colonoscopy videos with a whole duration of 5 hours. Expert colonoscopists were asked to identify all polyps in 9 de-identified colonoscopy videos, which were selected from archived video studies, with/without the benefit of the CNN overlay. Their findings were correlated with those of the CNN using CNN assisted expert review as the reference. The CNN identified polyps with an AUC of 0.99 and an accuracy of 96.4%. Indeed, in the analysis of colonoscopy videos involving the removal of 28 polyps, 4 expert reviewers identified 8 further (missed) polyps without CNN assistance and recognized an additional 17 polyps with CNN support. All polyps removed and recognized by the expert review were discovered by CNN, which showed a 7% false-positivity rate. This strategy could improve the ADR and lower interval cancers but it requires further studies to be adequately implemented.
AI can be used during endoscopic assessment to automatically recognize colorectal polyps and distinguish between malignant and non-malignant lesions. CAD is based on the latency time between the image acquisition to its processing for the ultimate visualization on the screen. This model was able to detect polyps with a 96.5% sensitivity[62,63].
A recent RCT estimated the impact of an automatic polyp detection system based on DL during real-time endoscopy. This study enrolling 1058 patients demonstrated that the AI system enhanced ADR of almost 10%[64].
A prospective study of 55 patients used a prototype of a novel automated polyp detection software (APDS) for automated image-based polyp detection and with overall real-time polyp detection of 75%[65]. Smaller polyp size and flat polyp morphology were associated with insufficient polyp detection by the APDS.
Aside from CADe machinery, CADx has been used for differentiating between adenomas and hyperplastic polyps.
Byrne et al[66] suggested the use of computerized image analysis to diminish the variability in endoscopic detection and histological prediction. This AI model was trained using endoscopic videos and was able to discriminate among diminutive adenomas and hyperplastic polyps with high accuracy. Additionally, it predicted histology with a 94% accuracy, 98% sensitivity, 83% specificity, a negative and positive predictive value of 97% and 90%, respectively.
Moreover, an AI-assisted image classifier, based on non-optical magnified endoscopic NBI, has been employed to predict the histology of isolated colonic lesions[67], following the evaluation of 3509 colonic lesions. The most prevalent histological types were tubular adenoma (47.6%), carcinoma with deep invasion (15.9%), carcinomas with superficial invasion (7.9%), hyperplastic polyps (14.3%), sessile serrated polyps (7.9%) and tubulovillous adenomas (6.6%). The sensitivity of hyperplastic and serrated polyps was 96.6%, although it was lower for tubular adenoma and cancer. When investigating only diminutive colonic polyps, the correlation of surveillance colonoscopy interval using AI image classifier and histology was 0.97. Moreover, this classifier also showed high accuracy (88.2%) in the prediction of carcinoma with deep invasion, which is not endoscopically curable, and the HNPV and accuracy for carcinoma with deep invasion also suggested that it can assist to select treatable lesions.
The same author assessed the use of AI-assisted image classifiers in determining the feasibility of ER of large colonic lesions based on non-magnified images. The independent testing set included 76 large colonic lesions that fulfilled the indications for endoscopic submucosal dissection. Overall, the trained AI image classifier showed a 88.2% sensitivity (95%CI: 84.7-91.1%) in differentiating endoscopically curable vs incurable lesions with a 77.9% specificity (95%CI: 70.3-84.4%) and 85.5% accuracy (95%CI: 82.4-88.3%). This study determined a high accuracy of the trained AI image classifier in predicting the feasibility of curative ER of large colonic lesions. While the progress of AI using CNN is great for the recognition of specific mucosal patterns and image classification, in the next future the prediction performance might outperform an expert endoscopist[68].
Hotta et al[69] aimed to validate the effectiveness of endocytoscopy (EC)-CAD in diagnosing malignant or non-malignant colorectal lesions, by comparing diagnostic ability between expert and non-expert endoscopists, by using web-based tests. A validation test was produced using endocytoscopic images of 100 small colorectal lesions (< 10 mm). Diagnostic accuracies and sensitivities of EB-01 and non-expert for stained endocytoscopic images were 98.0% vs 69.0%, showing a diagnostic accuracy and sensitivity significantly higher to non-expert endoscopists when diagnosing small colorectal lesions.
A single-group open-label prospective study assessed the performance of real-time EC-CAD on 791 consecutive patients undergoing colonoscopy and 23 endoscopists to differentiate neoplastic polyps (adenomas) requiring resection from non-neoplastic polyps not requiring treatment, potentially reducing cost[70]. The results revealed a 96.4% negative predictive value of CAD with stained mode in the best-case whereas 93.7% in the worst-case scenario. Wile by using NBI, 96.5%, and 95.2% in the best and worst-case scenario.
Another study developed an automatic quality control system (AQCS) and assessed a hypothetical improvement of polyp and adenoma detection in clinical practice based on deep CNN. The primary outcome of the study was to assess the ADR in the 308 AQCS and 315 control group patients. AQCS significantly increased the ADR than the control group. A significant improvement was similarly seen in the polyp detection rate and the mean number of polyps identified per-procedure[71].
Finally, in a study including 117 patients with stage IIA CRC after radical surgery, an ANN-based scoring system, based on the tumor molecular features, recognized those with a high, moderate, and low probability of survival at 10-year surveillance interval[72]. The 10-year overall survival rates were 16.7%, 62.9%, and 100% (P < 0.001), whereas the 10-year disease-free survival rates were 16.7%, 61.8%, and 98.8%, respectively. This study revealed that the scoring system for stage IIA CRC high-risk individuals for a more aggressive therapeutic approach.
DL distinguishes patients with a complete response to neoadjuvant chemoradiotherapy for locally advanced rectal cancer with an 80% accuracy. This technology support might allow to choose patients particularly benefitting the conservative treatment than complete surgical resection[73]. This is the first study using DL to predict total pathological response after neoadjuvant chemoradiotherapy in locally advanced rectal cancer.
AI could represent an essential diagnostic method for endoscopists and gastroenterologists for the patient's treatments tailoring and prediction of their clinical outcomes.
AI seems particularly valuable in gastrointestinal endoscopy, to improve the detection of premalignant lesions and malignant, or inflammatory lesions, gastrointestinal bleeding, and pancreaticobiliary diseases[74].
However, current limitations of AI include the lack of high-quality datasets for ML development. Moreover, a substantial evidence used to elaborate ML algorithms comes only from preclinical studies[74]. Potential selection biases cannot be excluded in such cases. In this setting, a rigorous validation of AI performance before its employment in daily clinical practice is necessary.
A real measure of AI accuracy, should include as a side effect in the performances overfitting and spectrum bias[75].
Overfitting occurs when a learning model tailors itself too much on the training dataset and predictions are not well generalized to new datasets[75,76]. This effect is in open contradiction with the problem-solving principle of Occam’s razor, which states that simpler theories have a higher quality of prediction[77]. In worst cases of AI algorithm application, underfitting can occur, obtaining models that cannot evidence accurately the underlying structure of the dataset, thus obtaining also bad predictivity model features[78].
On the other hand, spectrum bias happens when the dataset used for model development is not representative of the target population[75,79]. To avoid an overestimation of the accuracy and generalization, an external validation dataset collected in a way that minimizes the spectrum bias, should be guaranteed. Besides, well-designed multicenter observational studies, are required for a stronger validation.
Certainly, it is also noteworthy to acknowledge ethical issues since AI is not aware of the patient’s choices or legal liabilities. The privacy issues could be addressed using federated datasets that don’t involve centralized servers.
Future randomized studies could directly increase the overall value (quality vs cost) of the CNN by examining its effects on surveillance colonoscopy, endoscopic time, polyps and ADR, and pathology charges.
Since AI science is in progress, the current limitations must be considered as a future challenge, so actually they are inherited also in the medicine applications, including difficult predictability of situations characterized by some uncertainty.
In general, AI is revolutionizing the technology and impacting also other ethical aspects like human work replacement by machines, but this has always been an open question since the industrial revolution.
What can be done is to promote the mutual collaboration through gastrointestinal endoscopy applications, to reciprocally benefit from the achievements in both science fields.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Viswanath YKS S-Editor: Wang JL L-Editor: A E-Editor: Li X
| 1. | Russell S, Norvig P. Artificial Intelligence: A Modern Approach, Global Edition. 3rd editon. London: Pearson, 2016. |
| 2. | Colom R, Karama S, Jung RE, Haier RJ. Human intelligence and brain networks. Dialogues Clin Neurosci. 2010;12:489-501. [PubMed] |
| 3. | Goodfellow I, Bengio Y, Courville A. Deep Learning. Cambridge: The MIT Press, 2016. |
| 4. | Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 956] [Cited by in RCA: 963] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 5. | Dent J. Barrett's esophagus: A historical perspective, an update on core practicalities and predictions on future evolutions of management. J Gastroenterol Hepatol. 2011;26 Suppl 1:11-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | Sharma P, Bergman JJ, Goda K, Kato M, Messmann H, Alsop BR, Gupta N, Vennalaganti P, Hall M, Konda V, Koons A, Penner O, Goldblum JR, Waxman I. Development and Validation of a Classification System to Identify High-Grade Dysplasia and Esophageal Adenocarcinoma in Barrett's Esophagus Using Narrow-Band Imaging. Gastroenterology. 2016;150:591-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 161] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 7. | Phoa KN, Pouw RE, Bisschops R, Pech O, Ragunath K, Weusten BL, Schumacher B, Rembacken B, Meining A, Messmann H, Schoon EJ, Gossner L, Mannath J, Seldenrijk CA, Visser M, Lerut T, Seewald S, ten Kate FJ, Ell C, Neuhaus H, Bergman JJ. Multimodality endoscopic eradication for neoplastic Barrett oesophagus: results of an European multicentre study (EURO-II). Gut. 2016;65:555-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 172] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 8. | Shaheen NJ, Sharma P, Overholt BF, Wolfsen HC, Sampliner RE, Wang KK, Galanko JA, Bronner MP, Goldblum JR, Bennett AE, Jobe BA, Eisen GM, Fennerty MB, Hunter JG, Fleischer DE, Sharma VK, Hawes RH, Hoffman BJ, Rothstein RI, Gordon SR, Mashimo H, Chang KJ, Muthusamy VR, Edmundowicz SA, Spechler SJ, Siddiqui AA, Souza RF, Infantolino A, Falk GW, Kimmey MB, Madanick RD, Chak A, Lightdale CJ. Radiofrequency ablation in Barrett's esophagus with dysplasia. N Engl J Med. 2009;360:2277-2288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1146] [Cited by in RCA: 972] [Article Influence: 60.8] [Reference Citation Analysis (0)] |
| 9. | Johnston MH, Eastone JA, Horwhat JD, Cartledge J, Mathews JS, Foggy JR. Cryoablation of Barrett's esophagus: a pilot study. Gastrointest Endosc. 2005;62:842-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 122] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 10. | Overholt BF, Panjehpour M, Halberg DL. Photodynamic therapy for Barrett's esophagus with dysplasia and/or early stage carcinoma: long-term results. Gastrointest Endosc. 2003;58:183-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 211] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 11. | Sharma P, Brill J, Canto M, DeMarco D, Fennerty B, Gupta N, Laine L, Lieberman D, Lightdale C, Montgomery E, Odze R, Tokar J, Kochman M. White Paper AGA: Advanced Imaging in Barrett's Esophagus. Clin Gastroenterol Hepatol. 2015;13:2209-2218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Mendel R, Ebigbo A, Probst A, Messmann H, Palm C. Barrett’s Esophagus Analysis Using Convolutional Neural Networks. In: Bildverarbeitung für die Medizin. Springer, 2017: 80-85. [RCA] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Souza LA, Papa JP, Palm C, Messmann H. Computer-aided diagnosis using deep learning in the evaluation of early oesophageal adenocarcinoma. Gut. 2019;68:1143-1145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 14. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Prinz F, de Souza LA, Papa J, Palm C, Messmann H. Real-time use of artificial intelligence in the evaluation of cancer in Barrett's oesophagus. Gut. 2020;69:615-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 124] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 15. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 272] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 16. | Sharma P, Hawes RH, Bansal A, Gupta N, Curvers W, Rastogi A, Singh M, Hall M, Mathur SC, Wani SB, Hoffman B, Gaddam S, Fockens P, Bergman JJ. Standard endoscopy with random biopsies versus narrow band imaging targeted biopsies in Barrett's oesophagus: a prospective, international, randomised controlled trial. Gut. 2013;62:15-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 245] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 17. | Curvers WL, van Vilsteren FG, Baak LC, Böhmer C, Mallant-Hent RC, Naber AH, van Oijen A, Ponsioen CY, Scholten P, Schenk E, Schoon E, Seldenrijk CA, Meijer GA, ten Kate FJ, Bergman JJ. Endoscopic trimodal imaging versus standard video endoscopy for detection of early Barrett's neoplasia: a multicenter, randomized, crossover study in general practice. Gastrointest Endosc. 2011;73:195-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 18. | van der Sommen F, Zinger S, Curvers WL, Bisschops R, Pech O, Weusten BL, Bergman JJ, de With PH, Schoon EJ. Computer-aided detection of early neoplastic lesions in Barrett's esophagus. Endoscopy. 2016;48:617-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (2)] |
| 19. | de Groof J, van der Sommen F, van der Putten J, Struyvenberg MR, Zinger S, Curvers WL, Pech O, Meining A, Neuhaus H, Bisschops R, Schoon EJ, de With PH, Bergman JJ. The Argos project: The development of a computer-aided detection system to improve detection of Barrett's neoplasia on white light endoscopy. United European Gastroenterol J. 2019;7:538-547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 20. | Swager AF, de Groof AJ, Meijer SL, Weusten BL, Curvers WL, Bergman JJ. Feasibility of laser marking in Barrett's esophagus with volumetric laser endomicroscopy: first-in-man pilot study. Gastrointest Endosc. 2017;86:464-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 21. | Swager AF, Tearney GJ, Leggett CL, van Oijen MGH, Meijer SL, Weusten BL, Curvers WL, Bergman JJGHM. Identification of volumetric laser endomicroscopy features predictive for early neoplasia in Barrett's esophagus using high-quality histological correlation. Gastrointest Endosc. 2017;85:918-926.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 22. | Evans JA, Poneros JM, Bouma BE, Bressner J, Halpern EF, Shishkov M, Lauwers GY, Mino-Kenudson M, Nishioka NS, Tearney GJ. Optical coherence tomography to identify intramucosal carcinoma and high-grade dysplasia in Barrett's esophagus. Clin Gastroenterol Hepatol. 2006;4:38-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 37] [Reference Citation Analysis (0)] |
| 23. | Leggett CL, Gorospe EC, Chan DK, Muppa P, Owens V, Smyrk TC, Anderson M, Lutzke LS, Tearney G, Wang KK. Comparative diagnostic performance of volumetric laser endomicroscopy and confocal laser endomicroscopy in the detection of dysplasia associated with Barrett's esophagus. Gastrointest Endosc. 2016;83:880-888.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 24. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25540] [Article Influence: 1824.3] [Reference Citation Analysis (7)] |
| 25. | Okumura S, Yasuda T, Ichikawa H, Hiwa S, Yagi N, Hiroyasu T. Unsupervised Machine Learning Based Automatic Demarcation Line Drawing System on Nbi Images of Early GC. Gastroenterology. 2019;156:S-937. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Quang T, Schwarz RA, Dawsey SM, Tan MC, Patel K, Yu X, Wang G, Zhang F, Xu H, Anandasabapathy S, Richards-Kortum R. A tablet-interfaced high-resolution microendoscope with automated image interpretation for real-time evaluation of esophageal squamous cell neoplasia. Gastrointest Endosc. 2016;84:834-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 27. | Kodashima S, Fujishiro M, Takubo K, Kammori M, Nomura S, Kakushima N, Muraki Y, Goto O, Ono S, Kaminishi M, Omata M. Ex vivo pilot study using computed analysis of endo-cytoscopic images to differentiate normal and malignant squamous cell epithelia in the oesophagus. Dig Liver Dis. 2007;39:762-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Shin D, Protano MA, Polydorides AD, Dawsey SM, Pierce MC, Kim MK, Schwarz RA, Quang T, Parikh N, Bhutani MS, Zhang F, Wang G, Xue L, Wang X, Xu H, Anandasabapathy S, Richards-Kortum RR. Quantitative analysis of high-resolution microendoscopic images for diagnosis of esophageal squamous cell carcinoma. Clin Gastroenterol Hepatol. 2015;13:272-279.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 29. | Tokai Y, Yoshio T, Aoyama K, Horie Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Tsuchida T, Hirasawa T, Sakakibara Y, Yamada T, Yamaguchi S, Fujisaki J, Tada T. Application of artificial intelligence using convolutional neural networks in determining the invasion depth of esophageal squamous cell carcinoma. Esophagus. 2020;17:250-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 30. | Nakanishi H, Doyama H, Ishikawa H, Uedo N, Gotoda T, Kato M, Nagao S, Nagami Y, Aoyagi H, Imagawa A, Kodaira J, Mitsui S, Kobayashi N, Muto M, Takatori H, Abe T, Tsujii M, Watari J, Ishiyama S, Oda I, Ono H, Kaneko K, Yokoi C, Ueo T, Uchita K, Matsumoto K, Kanesaka T, Morita Y, Katsuki S, Nishikawa J, Inamura K, Kinjo T, Yamamoto K, Yoshimura D, Araki H, Kashida H, Hosokawa A, Mori H, Yamashita H, Motohashi O, Kobayashi K, Hirayama M, Kobayashi H, Endo M, Yamano H, Murakami K, Koike T, Hirasawa K, Miyaoka Y, Hamamoto H, Hikichi T, Hanabata N, Shimoda R, Hori S, Sato T, Kodashima S, Okada H, Mannami T, Yamamoto S, Niwa Y, Yashima K, Tanabe S, Satoh H, Sasaki F, Yamazato T, Ikeda Y, Nishisaki H, Nakagawa M, Matsuda A, Tamura F, Nishiyama H, Arita K, Kawasaki K, Hoppo K, Oka M, Ishihara S, Mukasa M, Minamino H, Yao K. Evaluation of an e-learning system for diagnosis of gastric lesions using magnifying narrow-band imaging: a multicenter randomized controlled study. Endoscopy. 2017;49:957-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 31. | Osawa H, Yamamoto H. Present and future status of flexible spectral imaging color enhancement and blue laser imaging technology. Dig Endosc. 2014;26 Suppl 1:105-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 109] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 32. | Osawa H, Yamamoto H, Miura Y, Yoshizawa M, Sunada K, Satoh K, Sugano K. Diagnosis of extent of early gastric cancer using flexible spectral imaging color enhancement. World J Gastrointest Endosc. 2012;4:356-361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 24] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 33. | Kimura-Tsuchiya R, Dohi O, Fujita Y, Yagi N, Majima A, Horii Y, Kitaichi T, Onozawa Y, Suzuki K, Tomie A, Okayama T, Yoshida N, Kamada K, Katada K, Uchiyama K, Ishikawa T, Takagi T, Handa O, Konishi H, Kishimoto M, Naito Y, Yanagisawa A, Itoh Y. Magnifying Endoscopy with Blue Laser Imaging Improves the Microstructure Visualization in Early Gastric Cancer: Comparison of Magnifying Endoscopy with Narrow-Band Imaging. Gastroenterol Res Pract. 2017;2017:8303046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 34. | Yoshifuku Y, Sanomura Y, Oka S, Kuroki K, Kurihara M, Mizumoto T, Urabe Y, Hiyama T, Tanaka S, Chayama K. Clinical Usefulness of the VS Classification System Using Magnifying Endoscopy with Blue Laser Imaging for Early Gastric Cancer. Gastroenterol Res Pract. 2017;2017:3649705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Wu L, Zhang J, Zhou W, An P, Shen L, Liu J, Jiang X, Huang X, Mu G, Wan X, Lv X, Gao J, Cui N, Hu S, Chen Y, Hu X, Li J, Chen D, Gong D, He X, Ding Q, Zhu X, Li S, Wei X, Li X, Wang X, Zhou J, Zhang M, Yu HG. Randomised controlled trial of WISENSE, a real-time quality improving system for monitoring blind spots during esophagogastroduodenoscopy. Gut. 2019;68:2161-2169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 214] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 36. | Miyaki R, Yoshida S, Tanaka S, Kominami Y, Sanomura Y, Matsuo T, Oka S, Raytchev B, Tamaki T, Koide T, Kaneda K, Yoshihara M, Chayama K. Quantitative identification of mucosal gastric cancer under magnifying endoscopy with flexible spectral imaging color enhancement. J Gastroenterol Hepatol. 2013;28:841-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 37. | Kanesaka T, Lee TC, Uedo N, Lin KP, Chen HZ, Lee JY, Wang HP, Chang HT. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest Endosc. 2018;87:1339-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 129] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 38. | Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, Ohnishi T, Fujishiro M, Matsuo K, Fujisaki J, Tada T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 426] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 39. | Zhu Y, Wang QC, Xu MD, Zhang Z, Cheng J, Zhong YS, Zhang YQ, Chen WF, Yao LQ, Zhou PH, Li QL. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc. 2019;89:806-815.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 231] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 40. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 4848] [Article Influence: 440.7] [Reference Citation Analysis (2)] |
| 41. | Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, Ye XS, Do IG, Liu S, Gong L, Fu J, Jin JG, Choi MG, Sohn TS, Lee JH, Bae JM, Kim ST, Park SH, Sohn I, Jung SH, Tan P, Chen R, Hardwick J, Kang WK, Ayers M, Hongyue D, Reinhard C, Loboda A, Kim S, Aggarwal A. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1071] [Cited by in RCA: 1578] [Article Influence: 157.8] [Reference Citation Analysis (0)] |
| 42. | Lee J, An JY, Choi MG, Park SH, Kim ST, Lee JH, Sohn TS, Bae JM, Kim S, Lee H, Min BH, Kim JJ, Jeong WK, Choi DI, Kim KM, Kang WK, Kim M, Seo SW. Deep Learning-Based Survival Analysis Identified Associations Between Molecular Subtype and Optimal Adjuvant Treatment of Patients With Gastric Cancer. JCO Clin Cancer Inform. 2018;2:1-14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 43. | Nilsaz-Dezfouli H, Abu-Bakar MR, Arasan J, Adam MB, Pourhoseingholi MA. Improving Gastric Cancer Outcome Prediction Using Single Time-Point Artificial Neural Network Models. Cancer Inform. 2017;16:1176935116686062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 44. | Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2805] [Cited by in RCA: 2739] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 45. | Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3126] [Cited by in RCA: 3183] [Article Influence: 132.6] [Reference Citation Analysis (0)] |
| 46. | Kato T, Yagi N, Kamada T, Shimbo T, Watanabe H, Ida K; Study Group for Establishing Endoscopic Diagnosis of Chronic Gastritis. Diagnosis of Helicobacter pylori infection in gastric mucosa by endoscopic features: a multicenter prospective study. Dig Endosc. 2013;25:508-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 103] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 47. | Zheng W, Zhang X, Kim JJ, Zhu X, Ye G, Ye B, Wang J, Luo S, Li J, Yu T, Liu J, Hu W, Si J. High Accuracy of Convolutional Neural Network for Evaluation of Helicobacter pylori Infection Based on Endoscopic Images: Preliminary Experience. Clin Transl Gastroenterol. 2019;10:e00109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 48. | Huang CR, Sheu BS, Chung PC, Yang HB. Computerized diagnosis of Helicobacter pylori infection and associated gastric inflammation from endoscopic images by refined feature selection using a neural network. Endoscopy. 2004;36:601-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 49. | Shichijo S, Nomura S, Aoyama K, Nishikawa Y, Miura M, Shinagawa T, Takiyama H, Tanimoto T, Ishihara S, Matsuo K, Tada T. Application of Convolutional Neural Networks in the Diagnosis of Helicobacter pylori Infection Based on Endoscopic Images. EBioMedicine. 2017;25:106-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 50. | Itoh T, Kawahira H, Nakashima H, Yata N. Deep learning analyzes Helicobacter pylori infection by upper gastrointestinal endoscopy images. Endosc Int Open. 2018;6:E139-E144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 130] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 51. | Nakashima H, Kawahira H, Kawachi H, Sakaki N. Artificial intelligence diagnosis of Helicobacter pylori infection using blue laser imaging-bright and linked color imaging: a single-center prospective study. Ann Gastroenterol. 2018;31:462-468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (1)] |
| 52. | Maida M, Macaluso FS, Ianiro G, Mangiola F, Sinagra E, Hold G, Maida C, Cammarota G, Gasbarrini A, Scarpulla G. Screening of colorectal cancer: present and future. Expert Rev Anticancer Ther. 2017;17:1131-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 125] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 53. | Winawer SJ, Zauber AG, Ho MN, O'Brien MJ, Gottlieb LS, Sternberg SS, Waye JD, Schapiro M, Bond JH, Panish JF. Prevention of colorectal cancer by colonoscopic polypectomy. The National Polyp Study Workgroup. N Engl J Med. 1993;329:1977-1981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3107] [Cited by in RCA: 3126] [Article Influence: 97.7] [Reference Citation Analysis (1)] |
| 54. | Patel SG, Ahnen DJ. Prevention of interval colorectal cancers: what every clinician needs to know. Clin Gastroenterol Hepatol. 2014;12:7-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 55. | Pohl H, Robertson DJ. Colorectal cancers detected after colonoscopy frequently result from missed lesions. Clin Gastroenterol Hepatol. 2010;8:858-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 203] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 56. | Corley DA, Levin TR, Doubeni CA. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:2541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 57. | Anderson JC, Butterly LF. Colonoscopy: quality indicators. Clin Transl Gastroenterol. 2015;6:e77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 58. | Maida M, Camilleri S, Manganaro M, Garufi S, Scarpulla G. New endoscopy advances to refine adenoma detection rate for colorectal cancer screening: None is the winner. World J Gastrointest Oncol. 2017;9:402-406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 59. | Bond A, Sarkar S. New technologies and techniques to improve adenoma detection in colonoscopy. World J Gastrointest Endosc. 2015;7:969-980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 60. | Waldmann E, Britto-Arias M, Gessl I, Heinze G, Salzl P, Sallinger D, Trauner M, Weiss W, Ferlitsch A, Ferlitsch M. Endoscopists with low adenoma detection rates benefit from high-definition endoscopy. Surg Endosc. 2015;29:466-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 61. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology. 2018;155:1069-1078.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 432] [Article Influence: 61.7] [Reference Citation Analysis (1)] |
| 62. | Misawa M, Kudo SE, Mori Y, Cho T, Kataoka S, Yamauchi A, Ogawa Y, Maeda Y, Takeda K, Ichimasa K, Nakamura H, Yagawa Y, Toyoshima N, Ogata N, Kudo T, Hisayuki T, Hayashi T, Wakamura K, Baba T, Ishida F, Itoh H, Roth H, Oda M, Mori K. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology. 2018;154:2027-2029.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 269] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 63. | Guizard N, Ghalehjegh SH, Henkel M, Ding L, Shahidi N, Jonathan GRP, Lahr RE, Chandelier F, Rex D, Byrne MF. Artificial Intelligence for Real-Time Multiple Polyp Detection with Identification, Tracking, and Optical Biopsy During Colonoscopy. Gastroenterology. 2019;156:S-48. [DOI] [Full Text] |
| 64. | Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, Li Y, Xu G, Tu M, Liu X. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 547] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 65. | Klare P, Sander C, Prinzen M, Haller B, Nowack S, Abdelhafez M, Poszler A, Brown H, Wilhelm D, Schmid RM, von Delius S, Wittenberg T. Automated polyp detection in the colorectum: a prospective study (with videos). Gastrointest Endosc. 2019;89:576-582.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 66. | Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 411] [Article Influence: 68.5] [Reference Citation Analysis (0)] |
| 67. | Lui TKL, Yee K, Wong K, Leung WK. Artificial Intelligence Image Classifier Based on Nonoptical Magnified Images Accurately Predicts Histology and Endoscopic Resectability of Different Colonic Lesions. Gastroenterology. 2019;156:S-48. [DOI] [Full Text] |
| 68. | Lui TKL, Wong KKY, Mak LLY, Ko MKL, Tsao SKK, Leung WK. Endoscopic prediction of deeply submucosal invasive carcinoma with use of artificial intelligence. Endosc Int Open. 2019;7:E514-E520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 69. | Hotta K, Kudo S, Mori Y, Ikematsu H, Saito Y, Ohtsuka K, Misawa M, Itoh H, Oda M, Mori K. Computer-Aided Diagnosis For Small Colorectal Lesions: A Multi-Center Validation “Endobrain Study” Designed To Obtain Regulatory Approval. Gastrointest Endosc. 2019;89:AB76. [DOI] [Full Text] |
| 70. | Mori Y, Kudo SE, Misawa M, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y, Maeda Y, Takeda K, Nakamura H, Ichimasa K, Kudo T, Hayashi T, Wakamura K, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 355] [Article Influence: 50.7] [Reference Citation Analysis (1)] |
| 71. | Su JR, Li Z, Shao XJ, Ji CR, Ji R, Zhou RC, Li GC, Liu GQ, He YS, Zuo XL, Li YQ. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: a prospective randomized controlled study (with videos). Gastrointest Endosc. 2020;91:415-424.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 208] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 72. | Peng JH, Fang YJ, Li CX, Ou QJ, Jiang W, Lu SX, Lu ZH, Li PX, Yun JP, Zhang RX, Pan ZZ, Wan de S. A scoring system based on artificial neural network for predicting 10-year survival in stage II A colon cancer patients after radical surgery. Oncotarget. 2016;7:22939-22947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 73. | Bibault JE, Giraud P, Housset M, Durdux C, Taieb J, Berger A, Coriat R, Chaussade S, Dousset B, Nordlinger B, Burgun A. Deep Learning and Radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci Rep. 2018;8:12611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 125] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 74. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology. 2020;158:76-94.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 323] [Article Influence: 64.6] [Reference Citation Analysis (1)] |
| 75. | Yang YJ, Bang CS. Application of artificial intelligence in gastroenterology. World J Gastroenterol. 2019;25:1666-1683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 211] [Cited by in RCA: 160] [Article Influence: 26.7] [Reference Citation Analysis (5)] |
| 76. | England JR, Cheng PM. Artificial Intelligence for Medical Image Analysis: A Guide for Authors and Reviewers. AJR Am J Roentgenol. 2019;212:513-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 77. | Schaffer J. What Not to Multiply Without Necessity. Australas J Philos. 2015;93:644–664. [DOI] [Full Text] |
| 78. | Everitt BS, Skrondal A. Cambridge Dictionary of Statistics. Cambridge: Cambridge University Press, 2010. |
| 79. | Park SH, Han K. Methodologic Guide for Evaluating Clinical Performance and Effect of Artificial Intelligence Technology for Medical Diagnosis and Prediction. Radiology. 2018;286:800-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 497] [Article Influence: 71.0] [Reference Citation Analysis (0)] |