Published online Jul 28, 2020. doi: 10.37126/aige.v1.i1.1
Peer-review started: June 1, 2020
First decision: June 18, 2020
Revised: July 14, 2020
Accepted: July 17, 2020
Article in press: July 17, 2020
Published online: July 28, 2020
Processing time: 51 Days and 19.3 Hours
The application of artificial intelligence (AI), especially machine learning or deep learning (DL), is advancing at a rapid pace. The need for increased accuracy at endoscopic visualisation of the gastrointestinal (GI) tract is also growing. Convolutional neural networks (CNNs) are one such model of DL, which have been used for endoscopic image analysis, whereby computer-aided detection and diagnosis of GI pathology can be carried out with increased scrupulousness. In this article, we briefly focus on the framework of the utilisation of CNNs in GI endoscopy along with a short review of a few published AI-based articles in the last 4 years.
Core tip: The convolutional neural network (CNN), a deep learning model, has gained immense success in endoscopy image analysis, with its application to diagnose and detect gastrointestinal (GI) pathology at endoscopy. This article shares a basic framework of the utilisation of CNNs in GI endoscopy, along with a concise review of a few published AI-based endoscopy articles in the last 4 years.
- Citation: Viswanath YK, Vaze S, Bird R. Application of convolutional neural networks for computer-aided detection and diagnosis in gastrointestinal pathology: A simplified exposition for an endoscopist. Artif Intell Gastrointest Endosc 2020; 1(1): 1-5
- URL: https://www.wjgnet.com/2689-7164/full/v1/i1/1.htm
- DOI: https://dx.doi.org/10.37126/aige.v1.i1.1
The role of artificial intelligence (AI), specifically machine learning (ML) or deep learning (DL), in medicine is evolving and studies have surfaced beholding its advantages in performing gastrointestinal (GI) endoscopy[1,2]. The pace of AI utilisation in medicine will further increase, especially in the coming years as a “new normal” is established post-coronavirus disease 2019 (COVID-19). Already, there is evidence of the advantages of AI utilisation in the diagnosis of various pathologies such as colonic polyps, esophagitis and GI cancer. It is also a fact that the translation of gained experience and skills over many years to a novice trainee is not easy and bound with initial problems, and raises errors whether in diagnosis or decision making. We believe that ML could play a resolute role in passing on this knowledge and facilitate better patient management.
Though computer programmes mimicking human cognitive functions have existed since the 1950s, it is only in the 1980s onwards that ML, followed by DL, applications have been studied in medical fields[3]. The future is looking likely to be increasingly automated and therefore driving AI research safely and fairly, with increased accuracy and interpretability, would reduce the dependency on skilled professionals, while concurrently aiding patient management at an early stage. There is increasing evidence that these results in a reduction in time-to-treatment and facilitate early patient management. However, AI in gastroenterology comes with some assurance as well as drawbacks.
Recent advances in AI as applied to medicine have largely come through ML, in which mathematical computer algorithms learn to interpret complex patterns in data. Specifically, DL, a subclass of ML originally inspired by the brain, uses layers of artificial neurons to form a “neural network” which maps inputs to an output. Typically, these networks are “trained” on large amounts of manually labelled data, in which example input-output pairs are provided to the model to enable it to “learn”. Of most interest to us in this article are the DL models which have achieved great success in image analysis tasks, namely convolutional neural networks (CNNs)[1-3]. We briefly focus on a high-level outline of the utilisation of CNNs in a simplified form to enable an endoscopist to cognize, along with a concise review of a few published GI endoscopy articles on AI in the last 4 years.
CNN's are a type of DL model, commonly used to analyse endoscopy images.
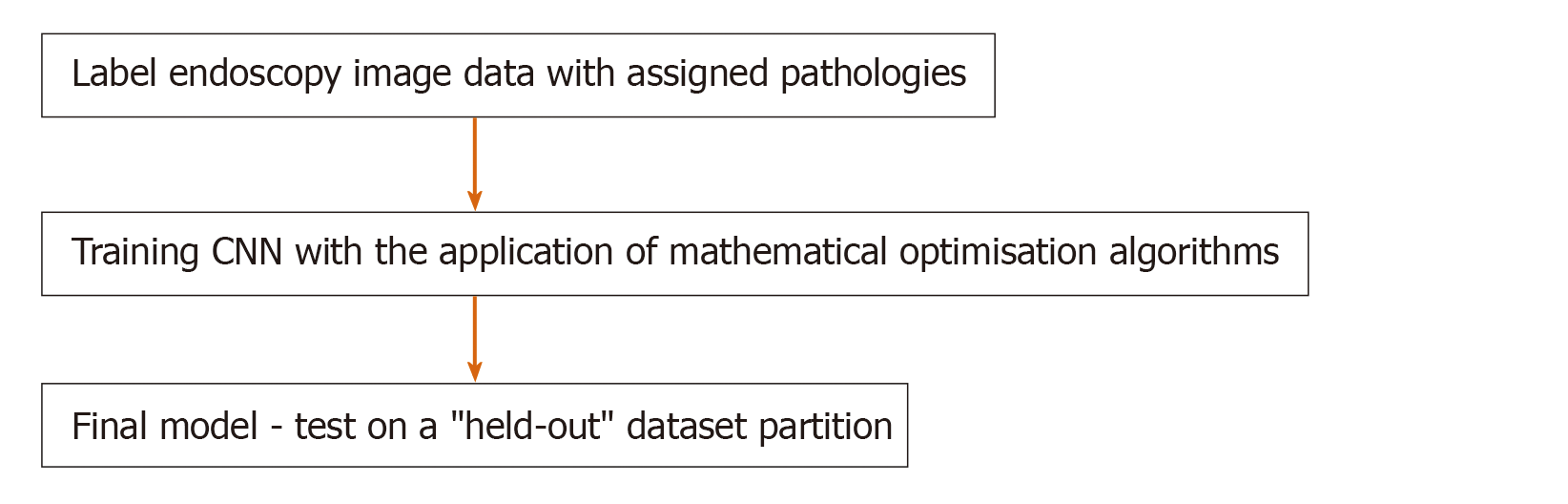
Figure 1 illustrates the CNN training method where ultrasound scan images have been used, highlighting the salient steps of the DL framework. At a high level, the CNN is a parametric model which maps an input - in this case, an image - to an output. The output can take a variety of forms: from a classification (a label of the image containing or not containing a tumour); to a detection (a bounding box around the tumour); to a segmentation (specification of exactly which pixels in the image contain the tumour)[4]. The model is “trained” by giving the model multiple (usually, thousands) of examples of input-output pairs.
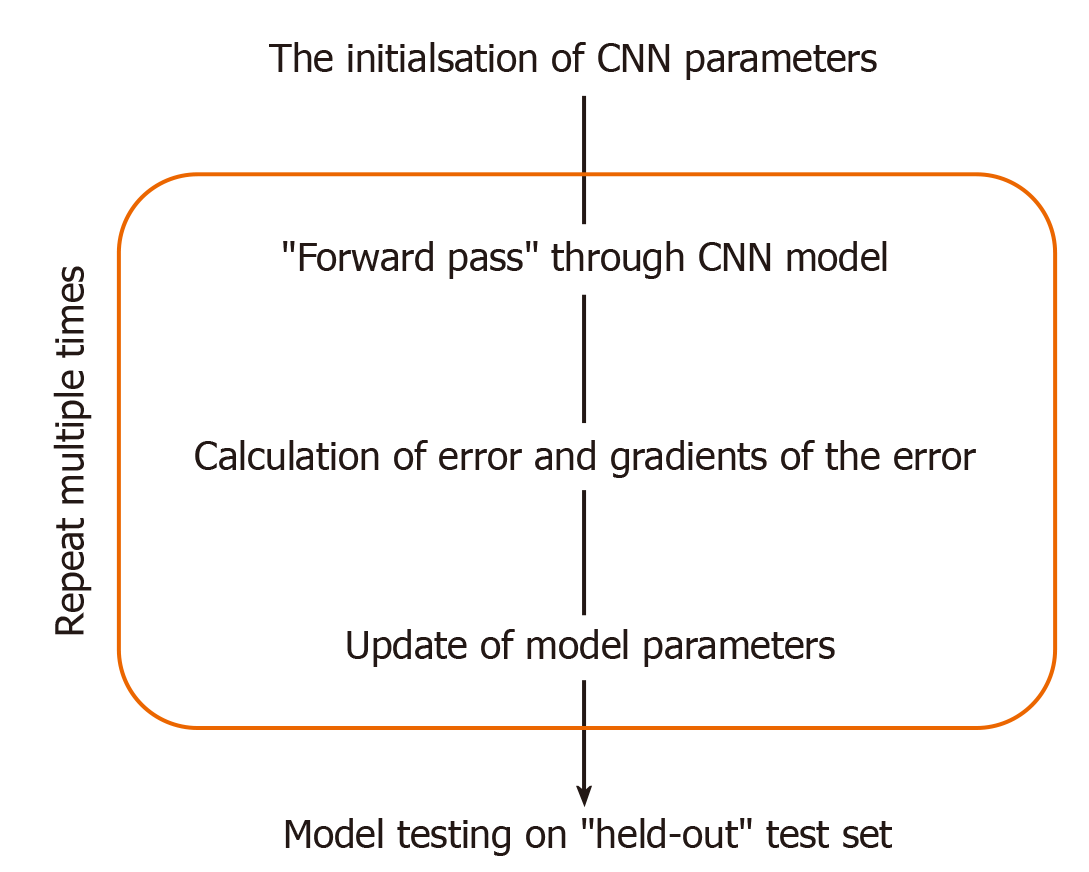
This training (Figure 2) involves, given an input image, computing the error between the model’s prediction and the manual label, with the parameters of the model then adjusted to reduce this error. This process is repeated numerous times until the performance of the model is acceptable, with its final accuracy computed on a held-out “test set“ of manually labelled images which it has not seen during training. Figure 1 illustrates the CNN framework, with the process rephrased in words in Figure 3. Figure 2 expands upon the training process specifically, in which the CNN parameters are iteratively updated so that its predictions are in closer alignment with the manual expert annotations. We highlight that the process involves partitioning the dataset into “training“ and “testing“ data, with the final model evaluation done on the “held-out“ test set - on images which the CNN has not seen during training. In this way, the model’s performance on this test set can be used to approximate how well the model generalises to images, not in the dataset.
The training has been well described in the recent article[2], where authors have highlighted splitting the computer learning model into training followed by validation set and then the test set. The initial game was to train the model to predict labelled image pathology followed by validation. This will allow the model to detect unseen pathology and lastly evaluate the outcomes of the trained model with optimal hyperparameters.
It can be seen, here, that by ensuring that the manual labels are generated by expert physicians, the model could encode this knowledge and be used to transfer it to trainees. The validation of this idea is an interesting avenue of research, in which two sets of labels could be collected for the “test-set“, from both experts and non-experts, to see if the CNN predictions can better align with the experts’ annotation than those of the less experienced physicians.
Data augmentation is one technique that can be used to supplement and amplify a small or limited dataset[2,4]. This can be beneficial in increasing data variability, thus exposing CNN to more examples to learn from and improve final model accuracy. Traditional augmentation methods on image data include image scaling and rotation, as well as manipulation of an images’ brightness, contrast or saturation. Synthesised images generated from a class of DL algorithms known as generative adversarial networks have also augmented data used to train CNNs, Frid-Adar et al[5].
We also highlight the potential risks of the naive implementation of this technology. The model will encode and operate well on patterns seen in the training data but will fail (often catastrophically and uninterpretably) when exposed to unseen patterns. As such, researchers and clinicians must carefully ascertain whether biases are encoded into the curated datasets. These biases may be clinical (e.g., omitted pathologies in the data) but also socio-technical (e.g. underrepresentation of sub-groups according to age, ethnicity, gender etc.). We direct the reader to further work on this topic[6].
The CNN training process has been well described in a recent article[2] where the authors have highlighted splitting the dataset into training, validation and test sets. The initial game was to train the model to predict labelled image pathology followed by validation.
The use of CNN’s in the early detection of oesophageal carcinoma has been published by Medel et al[7]. Here, the authors concluded improvement in sensitivity to 0.94 and specificity to 0.88. They defined C0 as a non-cancerous area and C1 as a cancerous area, highlighting the two regions and ultimately classified images as cancerous or non-cancerous using a patch-based approach. They concluded that future studies should include a greater number of images in the training set. Though adenoma detection rates at colonoscopy is variable with human interpretation, polyps’ localisation and detection rate using a CNN has been shown to improve accuracy to 96.4%. This, in turn, can affect a reduction in colorectal interval cancers and associated cancer mortality[8].
The CNN can be used as an image feature extractor along with a support vector machine (SVM) as an aid for polyp classification. Shape, size and surface characteristics guide the attending gastrointestinal physician to identify and differentiate benign and malignant polyps. The accuracy of detection and diagnosis is variable depending on the experience of the endoscopist and the equipment. It has been shown that AI-based systems increase the accuracy of diagnosis and detection rate of polyps.
A Japanese team published an article on polyp classification in 2017, where they used a CNN to extract features from the endoscopic image and an SVM to classify colonic polyps. The SVM algorithms are used primarily for classification and regression analysis. In this study, the authors noted increased accuracy by using multiple CNN-SVM classifiers[9]. A further improvement in the detection and classification can be achieved through improved extraction methods such as wavelet colour texture feature extraction. This is nicely illustrated in an article by Billlah et al[10]. In another study, authors showed an accuracy of 78.4% to differentiate adenomatous vs non-adenomatous colonic polyps[11]. The system used linked colour imaging and showed a sensitivity of 83% and specificity of 70.1%[9]. Likewise, AI has been used to classify inflammatory bowel disease with 90% accuracy[12]. In another study authors collected and tagged 6 colorectal segments from 100 patients - they inferred the computer-aided detection system has potential for automatic identification of persistent histological inflammation in patients with ulcerative colitis[13].
AI use in medicine is likely to rise fast along with its endoscopy applications, followed by a noticeable surge in investment by big industry players. Gastrointestinal physicians will witness many breakthroughs in the coming years; however, a lack of proper legislation and clinical governance structure needs to be addressed soon. This requires evidence-based consensus and acceptable international standards without compromising a patient’s safety in the coming years. Likewise, several technical issues within AI must be addressed, such as algorithm interpretability, fairness in results, and diverse representation in the dataset. However, reduction of errors due to endoscopist fatigue, inter-observer variability and learner endoscopist misconception are few rewards of AI; all these can no-doubt be leveraged to improve patient management. One cannot answer, whether, in the coming years, AI will replace humans in performing the endoscopies themselves! Indisputably, AI is here to stay and will play a vital role in the post-COVID-19 “new normal“ era.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United Kingdom
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): E
P-Reviewer: Hammoud CM, Inamdar S, Pang S, Kim GH S-Editor: Wang JL L-Editor: A E-Editor: Li JH
| 1. | Ruffle JK, Farmer AD, Aziz Q. Artificial Intelligence-Assisted Gastroenterology- Promises and Pitfalls. Am J Gastroenterol. 2019;114:422-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 2. | van der Sommen F, de Groof J, Struyvenberg M, van der Putten J, Boers T, Fockens K, Schoon EJ, Curvers W, de With P, Mori Y, Byrne M, Bergman JJGHM. Machine learning in GI endoscopy: practical guidance in how to interpret a novel field. Gut. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 3. | Min JK, Kwak MS, Cha JM. Overview of Deep Learning in Gastrointestinal Endoscopy. Gut Liver. 2019;13:388-393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 116] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 4. | Ragab DA, Sharkas M, Marshall S, Ren J. Breast cancer detection using deep convolutional neural networks and support vector machines. PeerJ. 2019;7:e6201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 5. | Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing. 2018;321:321-331. [RCA] [DOI] [Full Text] [Cited by in Crossref: 703] [Cited by in RCA: 554] [Article Influence: 79.1] [Reference Citation Analysis (0)] |
| 6. | Parikh RB, Teeple S, Navathe AS. Addressing Bias in Artificial Intelligence in Health Care. JAMA. 2019;322:2377-2378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 310] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 7. | Mendel R, Ebigbo A, Probst A, Messmann H, Palm C. Barrett’s Esophagus Analysis Using Convolutional Neural Networks. In: Bildverarbeitung für die Medizin. Springer, 2017: 80-85. [RCA] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 8. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology. 2018;155:1069-1078.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 432] [Article Influence: 61.7] [Reference Citation Analysis (1)] |
| 9. | Murata M, Usami H, Iwahori Y, Wang AL, Ogasawara N, Kasugai K. Polyp classification using multiple CNN-SVN classifiers from endoscopy images. In: Patterns 2017: the Ninth International Conferences on Pervasive Patterns and Applications. International Academy, Research, and Industry Association, 2017. Available from: https://www.thinkmind.org/download.php?articleid=patterns_2017_8_30_78008/. |
| 10. | Billah M, Waheed S. Gastrointestinal polyp detection in endoscopic images using an improved feature extraction method. Biomed Eng Lett. 2018;8:69-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 11. | Min M, Su S, He W, Bi Y, Ma Z, Liu Y. Computer-aided diagnosis of colorectal polyps using linked color imaging colonoscopy to predict histology. Sci Rep. 2019;9:2881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 12. | Mossotto E, Ashton JJ, Coelho T, Beattie RM, MacArthur BD, Ennis S. Classification of Paediatric Inflammatory Bowel Disease using Machine Learning. Sci Rep. 2017;7:2427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 13. | Maeda Y, Kudo SE, Mori Y, Misawa M, Ogata N, Sasanuma S, Wakamura K, Oda M, Mori K, Ohtsuka K. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc. 2019;89:408-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 164] [Article Influence: 27.3] [Reference Citation Analysis (0)] |