Published online Sep 28, 2024. doi: 10.35711/aimi.v5.i1.93993
Revised: August 27, 2024
Accepted: September 5, 2024
Published online: September 28, 2024
Processing time: 200 Days and 23.4 Hours
The presence of perineural invasion (PNI) in patients with rectal cancer (RC) is associated with significantly poorer outcomes. However, traditional diagnostic modalities have many limitations.
To develop a deep learning radiomics stacking nomogram model to predict preoperative PNI status in patients with RC.
We recruited 303 RC patients and separated them into the training (n = 242) and test (n = 61) datasets on an 8: 2 scale. A substantial number of deep learning and hand-crafted radiomics features of primary tumors were extracted from the arterial and venous phases of computed tomography (CT) images. Four machine learning models were used to predict PNI status in RC patients: support vector machine, k-nearest neighbor, logistic regression, and multilayer perceptron. The stacking nomogram was created by combining optimal machine learning models for the arterial and venous phases with predicting clinical variables.
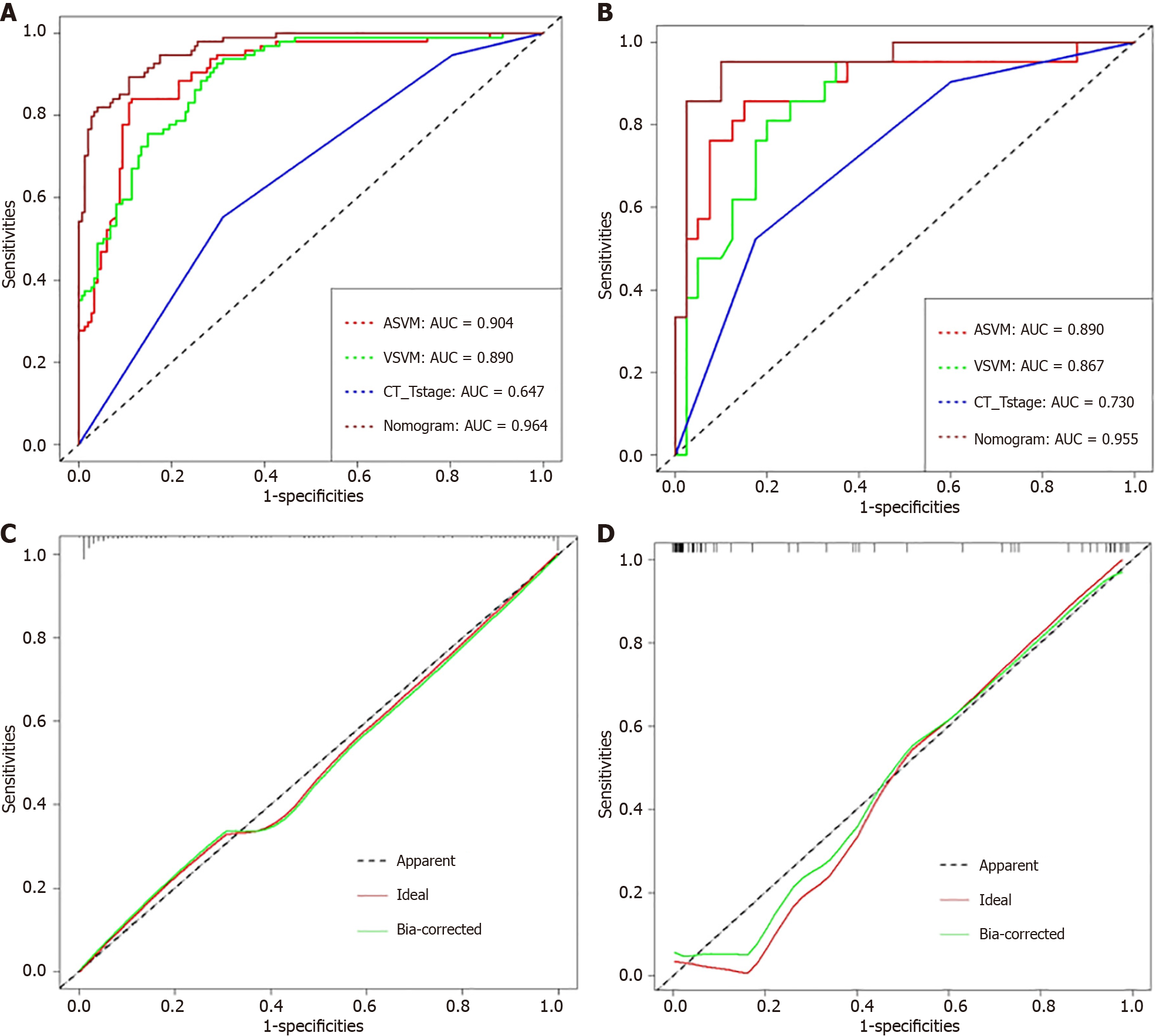
With an area under the curve (AUC) of 0.964 [95% confidence interval (CI): 0.944-0.983] in the training dataset and an AUC of 0.955 (95%CI: 0.900-0.999) in the test dataset, the stacking nomogram demonstrated strong performance in predicting PNI status. In the training dataset, the AUC of the stacking nomogram was greater than that of the arterial support vector machine (ASVM), venous SVM, and CT-T stage models (P < 0.05). Although the AUC of the stacking nomogram was greater than that of the ASVM in the test dataset, the difference was not particularly noticeable (P = 0.05137).
The developed deep learning radiomics stacking nomogram was effective in predicting preoperative PNI status in RC patients.
Core Tip: Four machine models (support vector machine, k-nearest neighbor, multilayer perceptron, and logistic regression) were used to predict the preoperative rectal cancer (RC) presence of perineural invasion (PNI) status, with good performance in both the arterial and venous phases. With an area under the curve of 0.964 in the training dataset and 0.955 in the test dataset, the stacking nomogram model to predict pretreatment PNI status had high predictive power and clinical utility, which can help diagnostic and treatment decision-making. Deep learning radiomics stacking models are rare in our RC PNI, which was also an innovation in our research.
- Citation: Zhao ZC, Liu JX, Sun LL. Preoperative perineural invasion in rectal cancer based on deep learning radiomics stacking nomogram: A retrospective study. Artif Intell Med Imaging 2024; 5(1): 93993
- URL: https://www.wjgnet.com/2644-3260/full/v5/i1/93993.htm
- DOI: https://dx.doi.org/10.35711/aimi.v5.i1.93993
Rectal cancer (RC) is the most common type of gastrointestinal cancer worldwide, and its incidence and mortality are steadily increasing, posing a severe threat to human health[1]. The two most distinguishing biological activities of malignant tumors are invasion and metastasis. Oncologists and physicians are becoming more aware of neural invasion in addition to the usual direct invasion, lymph node metastasis, and hematogenous metastasis.
One obvious method by which cancer cells spread is through neural invasion, also known as perineural invasion (PNI), which is the invasion of tumor cells around or through nerve fibers[2]. PNI is present in many different tumor types, including pancreatic ductal adenocarcinoma, gastric cancer, colorectal cancer, and prostate cancer. It plays a significant role in determining the pathological features and prognosis of malignant tumors by foretelling a high incidence of metastatic tumors, poor prognosis, and high rate of local recurrence[2,3]. PNI has a significant role in deciding whether patients benefit from postoperative chemotherapy and neoadjuvant chemoradiation[4-6]. Furthermore, it significantly affects the prognosis of individuals with rectal cancer who will survive over the long term. Therefore, physicians can benefit from knowing PNI status beforehand.
Traditional radiological methods, such as computed tomography (CT) and magnetic resonance imaging (MRI), do not determine the PNI status of rectal cancer. However, because RC is a temporally and spatially heterogeneous disease, the risk of invasive sampling and potential complications limit its application in tumor progression and real-time monitoring. As a result, a simple and noninvasive strategy to provide this critical information before clinicians make clinical treatment decisions must be developed.
Radiomics, which uses a large number of objective and quantitative imaging features to select imaging markers that are most closely related to clinical, pathological, molecular, and genetic characteristics, and then uses machine learning and statistical modeling to perform further quantitative analysis and analyze the correlation with clinical features, can noninvasively reflect tumor heterogeneity[7-9]. Several recent studies[10] have demonstrated that radiomics is a superior method for predicting PNI status in colorectal cancer. Guo et al[10] created a nomogram based on CT score and T2-weighted imaging score to predict PNI status in RC, and it performed the best [training set, area under the curve (AUC) = 0.906; test set, AUC = 0.884][11]. The results of the study demonstrated that radiomics can supplement conventional imaging techniques and aid physicians in decision-making. Additionally, a type of deep learning neural network that learns from the data itself is the convolutional neural network (CNN). Convolution is its central layer and is mostly utilized for segmentation, classification, and image recognition. Large data sets can be processed and the outcome of data analysis can be predicted and classified[12]. It is rarely stated that deep learning radiomics can be used to predict PNI status in RC.
Therefore, in this study, radiomics features of arterial and venous phases were extracted from enhanced CT images of patients with RC, and a deep learning radiomics nomogram was constructed to explore its application the prediction of PNI.
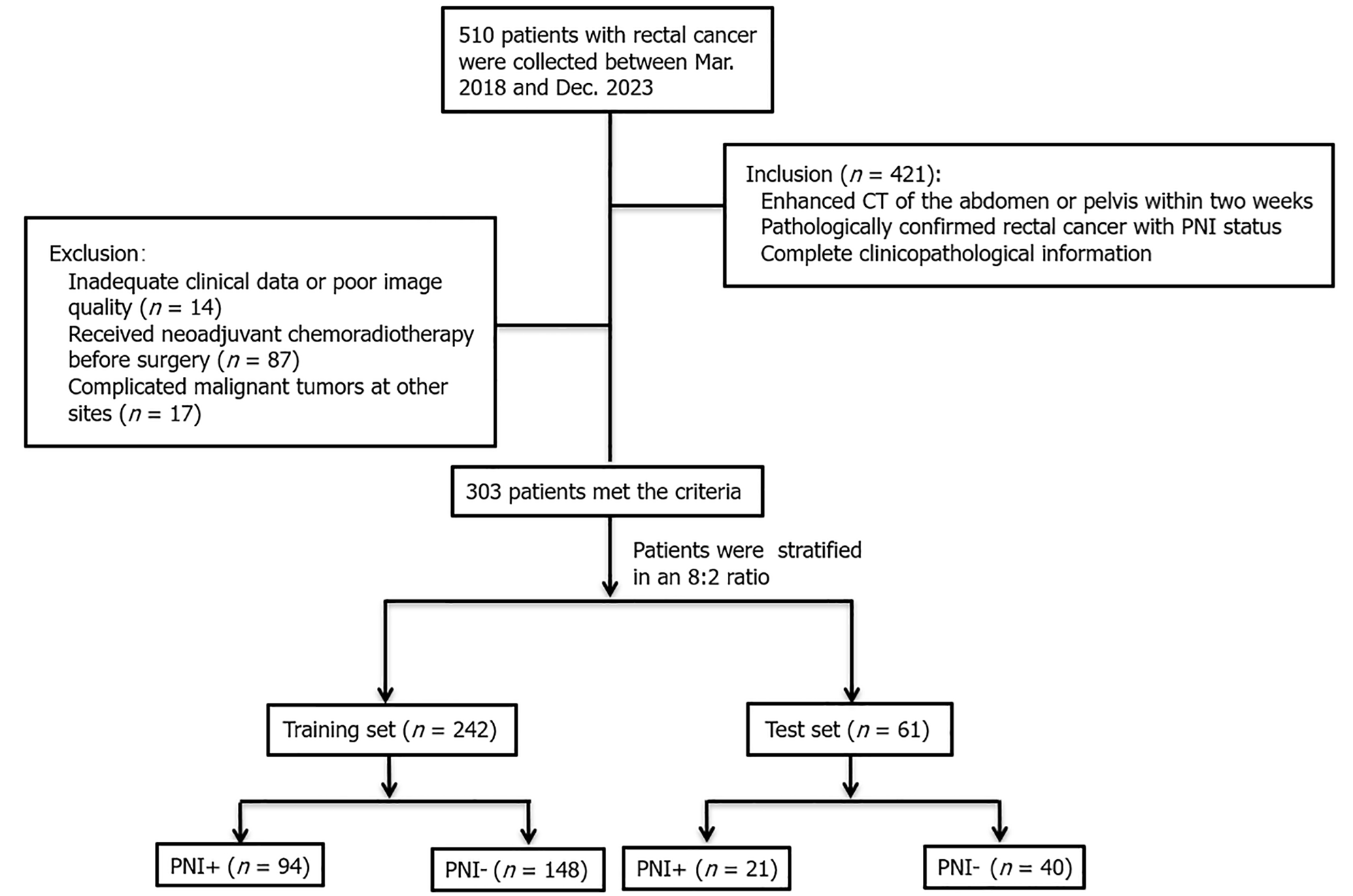
Patients with RC who underwent enhanced CT examination at our hospital between March 2018 and December 2023 were included in this study retrospectively. The following were the inclusion criteria for patients (Figure 1): (1) Pathologically confirmed RC with PNI status; (2) Within 2 wk prior to surgery, an enhanced abdominal CT scan was conducted; and (3) All clinical information and pertinent laboratory results were recorded, including age, sex, history of alcohol consumption and smoking, carcinoembryonic antigen (CEA), carbohydrate antigen (CA) 19-9, routine blood tests, blood lipids, and T stage of the CT report. Exclusion criteria were (Figure 1): (1) Neoadjuvant chemoradiotherapy prior to surgery; (2) Complicated malignant tumors at other sites; and (3) Inadequate clinical data or poor image quality. Ultimately, the study comprised 303 patients (mean age 65.94 ± 10.76 years, age range 24-91 years; 165 male and 117 female). The patients were assigned to training (n = 242) and test (n = 61) datasets.
Spiral CT (Philips iCT 256) showed that all patients had enhanced abdominal scans with the following settings: Matrix 512 × 512, transverse fault thickness of 5 mm, pitch 0.5 s, tube voltage 120 kV, and tube current autoregulation. Patients in the supine position were injected with 80-100 mL (300 mg mL) at a rate of 3.0 mL/s with a delay of 30-35 s and 60-70 s, resulting in arterial and venous phase images.
Two seasoned radiologists who were blind to all clinical and pathological data evaluated the CT reported T stage (CT-T stage) using CT-enhanced images (Table 1).
| Characteristics | Training set (n = 242) | P value | Test set (n = 61) | P value | ||
| PNI (n = 148) | PNI+ (n = 94) | PNI (n = 40) | PNI+ (n = 21) | |||
| Age, (mean ± SD) (years) | 67.01 ± 10.57 | 64.26 ± 12.06 | 0.063 | 67.33 ± 8.18 | 63.104 ± 8.66 | 0.065 |
| Gender | 0.248 | 0.86 | ||||
| Male | 85 (58.2) | 54 (39.1) | 20 (64.5) | 11 (35.5) | ||
| Female | 63 (65.6) | 33 (34.4) | 20 (66.7) | 10 (33.3) | ||
| Smoking | 0.882 | 0.396 | ||||
| No | 101 (60.8) | 65 (39.2) | 29 (69.0) | 13 (31.0) | ||
| Yes | 47 (51.8) | 29 (38.2) | 11 (57.9) | 8 (42.1) | ||
| HGB (g/L) | 126.98 ± 20.67 | 130.69 ± 20.00 | 0.071 | 128.881 ± 14.16 | 128.67 ± 22.28 | 0.965 |
| RBC (1012/L) | 4.28 ± 0.62 | 4.40 ± 0.44 | 0.126 | 4.40 ± 0.47 | 4.28 ± 0.48 | 0.339 |
| WBC (109/L) | 6.55 ± 1.79 | 6.84 ± 2.19 | 0.265 | 6.18 ± 1.68 | 6.54 ± 1.67 | 0.391 |
| PLT (109/L) | 229.14 ± 77.31 | 240.92 ± 76.73 | 0.248 | 231.05 ± 70.18 | 231.62 ± 50.56 | 0.974 |
| Lymphocyte(109/L) | 1.61 ± 0.59 | 1.63 ± 0.68 | 0.808 | 1.59 ± 0.68 | 1.71 ± 0.95 | 0.567 |
| Monocyte(109/L) | 0.46 ± 0.23 | 0.47 ± 017 | 0.667 | 0.40 ± 0.14 | 0.70 ± 1.02 | 0.1 |
| Neutrophil(109/L) | 4.27 ± 1.55 | 4.54 ± 1.94 | 0.243 | 3.96 ± 1.51 | 4.47 ± 1.70 | 0.228 |
| TG | 1.47 ± 1.11 | 1.31 ± 0.60 | 0.18 | 1.43 ± 0.72 | 1.59 ± 0.98 | 0.502 |
| Cholesterol | 4.58 ± 0.88 | 4.72 ± 0.98 | 0.232 | 4.82 ± 0.89 | 4.91 ± 1.08 | 0.707 |
| HDL | 1.12 ± 0.28 | 1.19 ± 0.32 | 0.088 | 1.42 ± 1.51 | 1.13 ± 0.24 | 0.392 |
| LDL | 2.79 ± 0.69 | 2.93 ± 0.85 | 0.164 | 2.96 ± 0.84 | 3.18 ± 1.09 | 0.392 |
| AproA | 1.24 ± 0.19 | 1.26 ± 0.20 | 0.406 | 1.29 ± 0.17 | 1.22 ± 0.15 | 0.126 |
| AproB | 0.89 ± 0.19 | 0.90 ± 0.22 | 0.795 | 0.94 ± 0.18 | 0.89 ± 0.19 | 0.279 |
| CEA (≥ 5 ng/mL) | 0.016 | 0.173 | ||||
| No | 94 (67.6) | 45 (32.4) | 28 (71.8) | 11 (28.2) | ||
| Yes | 54 (52.4) | 49 (47.6) | 12 (54.5) | 10 (45.5) | ||
| CA19-9 (≥ 37 U/mL) | 0.003 | 0.052 | ||||
| No | 136 (64.8) | 74 (35.2) | 37 (71.2) | 15 (28.8) | ||
| Yes | 12 (37.5) | 20 (62.5) | 3 (33.3) | 6 (66.7) | ||
| CT T stage | 0.000 | 0.006 | ||||
| 1/2 | 29 (85.3) | 5 (14.7) | 16 (88.6) | 2 (11.1) | ||
| 3 | 73 (66.4) | 37 (33.6) | 17 (68.0) | 8 (32.0) | ||
| 4 | 46 (46.9) | 52 (53.1) | 7 (38.9) | 11 (61.1) | ||
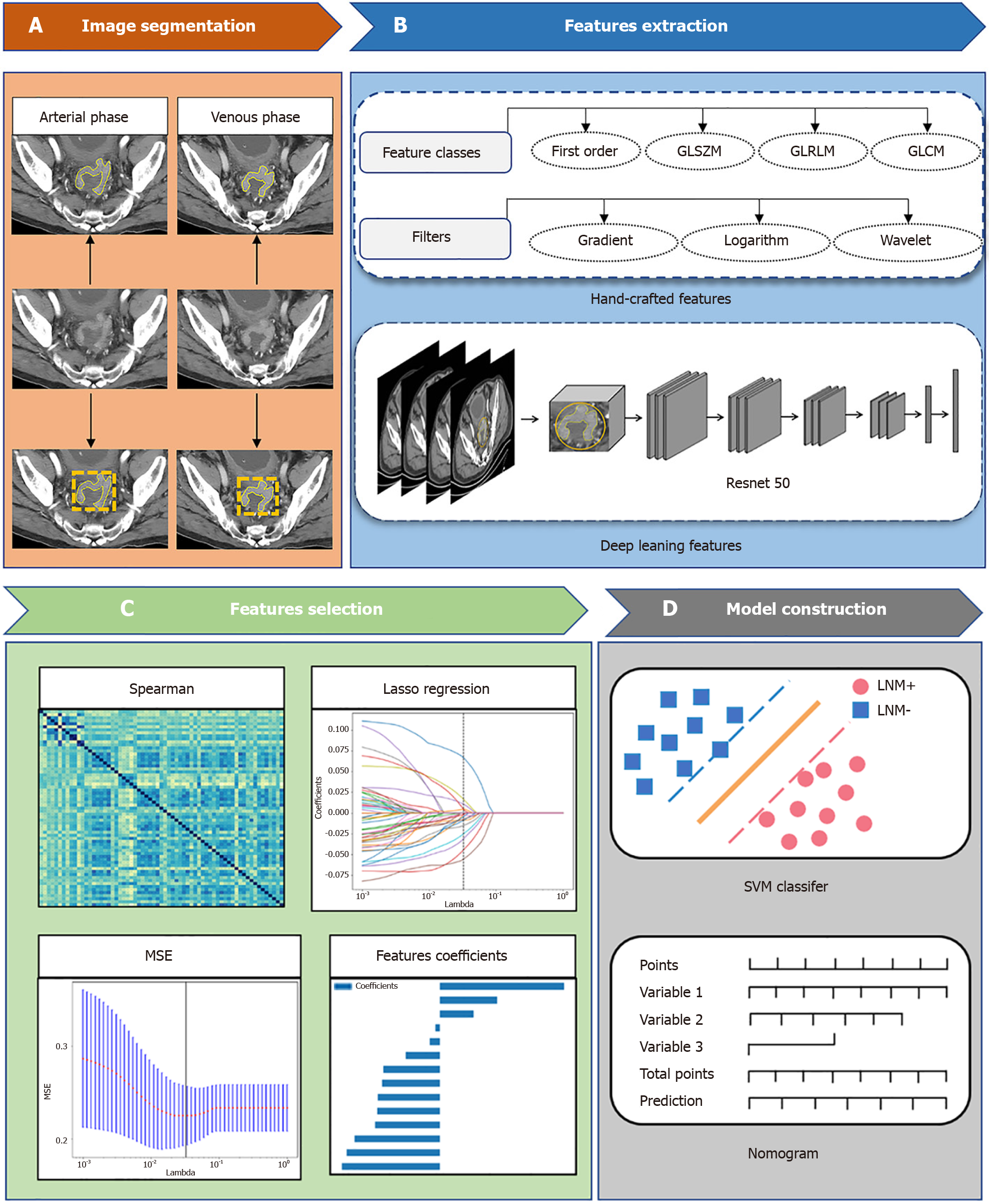
Using the open-source 3D Slicer software (www.3D-Slicer.com, version 4.13.2), two radiologists manually identified and separated main tumors from axial CT scans at arterial and venous stages. The pixel intensity was normalized to transform the images to standardized inputs, which had the intensity range from 1024 to 1024 HU and the unified abdominal window (window level 50 and window width 350). The two radiologists repeated manual segmentation on the same group of 50 CT images to test the consistency of the two, intraclass correlation coefficients (CCs) used for consistency within the tester. For intraclass CCs used for consistency between examiner and assessors, only intraclass CCs > 0.75 indicated that acceptable stability of the construction model. Regions of interest (ROIs) include tumor and necrosis, bleeding areas and avoid the use of intestinal gases and contents. Fir every patient, two ROIs (venous and arterial phases) were created.
CT images were resampled to the voxel size of 1 mm × 1 mm × 1 mm. The raw images were processed using log, exponential, square, square root, gradient, and high wavelet transform and low wavelet filter. First-order features (n = 342), shape features (n = 14), gray-level dependence matrix (n = 266), gray-level size zone matrix (GLSZM, n = 304), and gray-level run-length matrix (n = 306) were among the radiomics features.
For the extraction of deep learning features, we initialized deep CNNs (DCNNs) using the pretrained weights in ImageNet, and selected the maximum cross-sectional area and its upper and lower images as three-channel images. The CT images were cropped using a rectangular ROI around the tumor contours. The size of the tumor patch was adjusted to 224224 to meet the input size requirements of the pretrained CNN model. We used the same normalization technique as in the ImageNet dataset, subtracting the mean (0.485, 0.456 and 0.406) and dividing by the standard deviation (0.229, 0.224 and 0.225) to ensure that the input features of the image agreed with the mean and standard deviation during ImageNet training.
We constructed two different deep learning models (DCNNs) for the deep learning feature extraction of tumor ROI in the arterial and venous stage CT images, respectively. The DCNN model was based on the Resnet-50 backbone, extracted deep learning features for classification, and predicted the RC PNI status based on a large number of 2D patches extracted from the ROI of the main cohort. The largest cross-sectional area and its upper and lower cross-section lesions were selected from the arterial and venous stages of the ROI as the input model, and the CT slices of the extracted features were input into the hierarchical convolution structure of the DCNN, using its CNN structure and learning weights on ImageNet, to obtain accurate ROI features in the average pool layer of the DCNN using the ResNet50 architecture. Using the average pool layer of ResNet-50 as the output of feature extraction generated a fixed size feature vector (usually 2048 dimensions), which provided a uniform and stable input feature for subsequent classification tasks, reducing the variability brought by different ROI sizes. The feature representation of this layer summarized the information about the entire ROI, providing a fixed-length feature vector suitable for subsequent classification tasks. We also froze most or all of the convolutional layers, only fine-tuning them in the final fully connected layer. This reduced the training time while maintaining the stability of the pretraining features.
Following the segmented ROI, 3696 features were extracted from each arterial and venous phase ROI, consisting of 2048 deep learning features and 1648 radiomics features, respectively. All features were normalized to a standard numerical range. We assessed the stability of two radiologists' tumor delineation using the interclass CC)/intraclass CC in order to remove unstable features and keep features with intraclass CC > 0.75. The Mann–Whitney U test (P < 0.05) eliminated the duplicate features. The features with the highest correlation with the outcome but the lowest correlation among the features were chosen using the maximum correlation and the minimum redundancy (mRMR). We selected the lambda value that yielded the least amount of error as the final parameter. LASSO regression lowered the coefficient of 0 and prevented overfitting and multicollinearity of the model when combined with fivefold cross-validation. The study flow chart, which includes image preprocessing, feature extraction, feature selection, and model building, is depicted in Figure 2.
Preoperative PNI status was developed in the radiomics models (arterial and venous phase) using four machine learning models: Support vector machine (SVM), logistic regression (LR), multilayer perceptron (MLP), and k-nearest neighbor (KNN). The effectiveness of these four machine learning models in determining PNI status was assessed using the AUC and receiver operating characteristic (ROC) curves. We chose the most effective machine learning model by using the DeLong test to determine whether there were significant differences between the four distinct ROC curves.
For continuous clinical factors (age, blood routine index, blood lipid items, etc.) that differed between PNI+ and PNI groups, we used a t test. For categorical variables (gender, CA19-9, CEA level and CT-T stage), we utilized χ2 testing. Multivariate logistic regression was used to examine clinical characteristics with P < 0.05 to identify independent clinical predictors. Ultimately, the most effective arterial and venous phase machine learning models were combined with clinically predictive features to create a stacking nomogram. The DeLong test examined whether there were differences between stacking nomogram, clinical models and the ROC curves of the machine learning models. Hosmer–Lemeshow examined the nomogram for fit, and the model fitted well at P > 0.05. The agreement between the actual and anticipated values of the superimposed nomograms was evaluated using calibration curves. The decision curve was employed to evaluate the clinical net-gain of the nomogram.
Anaconda (https://www.anaconda.com/python3.7) and R (https://www.r-project.org/version4.1.2) were used for statistical analysis. When comparing the differences between training and test groups for continuous clinical variables, an independent sample t test was used if a normal distribution was satisfied; if not, the Mann–Whitney U test was utilized to assess the differences. To evaluate the differences between categorical clinical variables, we used Fisher's exact test or χ2 test. Ultimately, we used multiple logistic regression analysis to identify independent predictors, and two-sided P < 0.05 was considered statistically significant.
Age, gender, history of smoking, history of alcohol consumption, CA19-9 level, CEA level, routine blood index, and six blood lipid items did not significantly differ between the PNI and PNI+ groups in the training and test datasets (P > 0.05). CT T-stage was shown to be significant in the training and test datasets (P < 0.05). Clinical characteristics of the patients are shown in Table 1.
We extracted 3254 radiomic features from each ROI during the arterial and venous phases. Intraclass CC < 0.75 excluded 83 arterial phase features and 94 venous phase features. Then, 131 and 154 features were selected using the Mann–Whitney U test. The top 50 features were retained by mRMR, and the last 15 arterial phase and 13 venous phase predictive features were determined by LASSO regression combined with cross-validation.
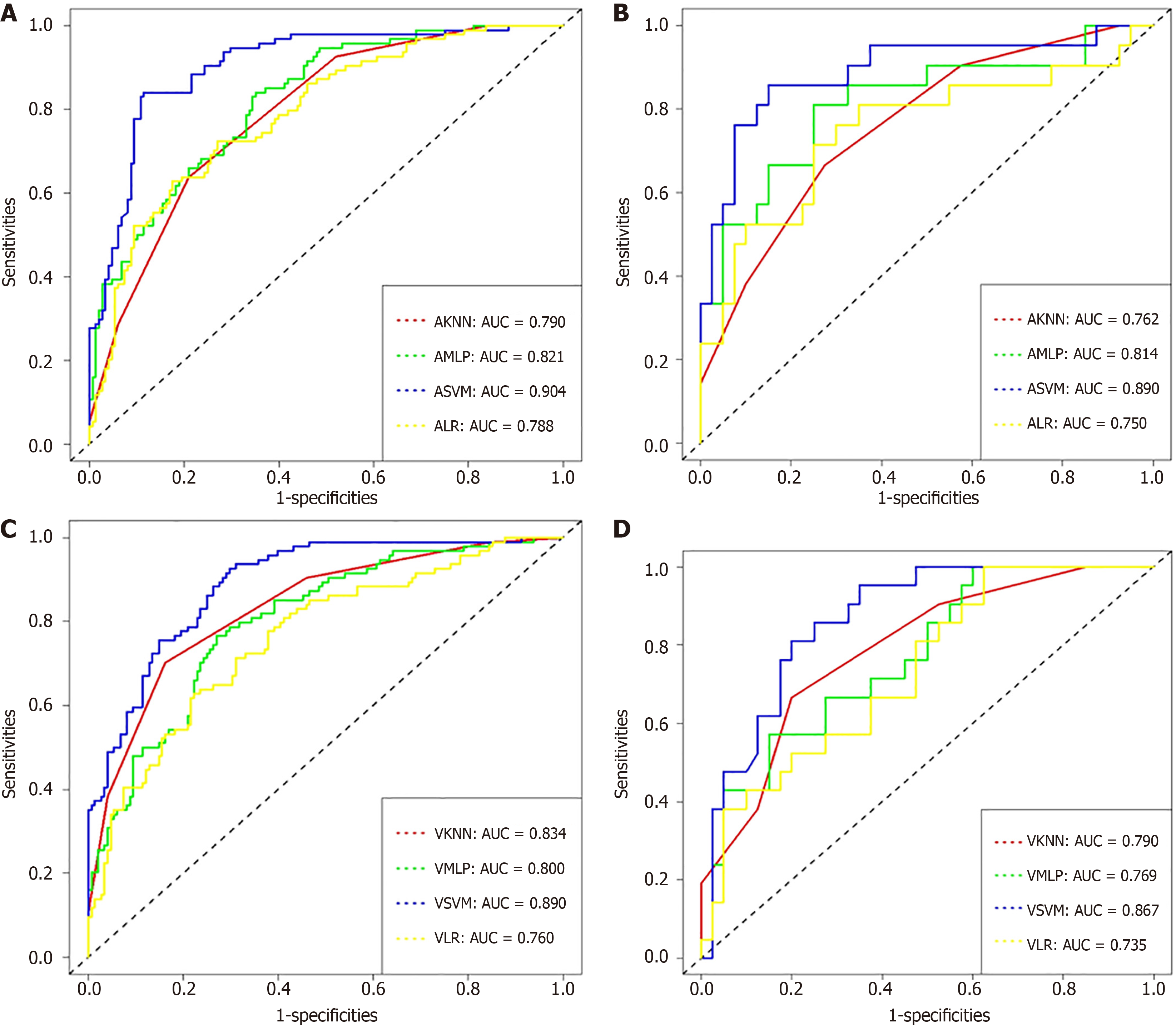
Figure 3 and Table 2 show the ROC curves and AUCs for the four machine learning models. The established radiomics model accurately predicted the preoperative PNI status in RC patients, according to the AUCs of the four machine learning models. The results showed that arterial SVM (ASVM) and venous SVM (VSVM) had AUCs of 0.904 and 0.890 in the training set, sensitivity of 0.840 and 0.926, and specificity of 0.885 and 0.703, respectively. SVM was the most effective model for the venous and arterial phases. In the test set, the AUC, sensitivity, and specificity of ASVM and VSVM were 0.890 and 0.867, 0.857 and 0.810, and 0.850 and 0.800, respectively. In the training group, the SVM models significantly outperformed the KNN, LR, and MLP models (P < 0.05). In the test group, however, the difference between the AUC of the ASVM model and the MLP model was not significant (P = 0.05938), nor was the difference between the AUC of the VSVM model and the KNN model (P = 0.15586).
| Classifiers | Training set | Test set | ||||||
| AUC | 95%CI | Sensitivity | Specificity | AUC | 95%CI | Sensitivity | Specificity | |
| ASVM | 0.904 | 0.865-0.943 | 0.840 | 0.885 | 0.890 | 0.794-0.987 | 0.857 | 0.850 |
| AKNN | 0.790 | 0.736-0.844 | 0.638 | 0.791 | 0.762 | 0.640-0.884 | 0.667 | 0.725 |
| AMLP | 0.821 | 0.769-0.873 | 0.840 | 0.649 | 0.814 | 0.691-0.937 | 0.810 | 0.750 |
| ALR | 0.788 | 0.731-0.846 | 0.723 | 0.730 | 0.750 | 0.607-0.893 | 0.714 | 0.750 |
| VSVM | 0.890 | 0.850-0.930 | 0.926 | 0.703 | 0.867 | 0.778-0.956 | 0.810 | 0.800 |
| VKNN | 0.834 | 0.783-0.884 | 0.702 | 0.838 | 0.790 | 0.676-0.904 | 0.667 | 0.800 |
| VMLP | 0.800 | 0.744-0.856 | 0.766 | 0.730 | 0.769 | 0.648-0.890 | 0.571 | 0.850 |
| VLR | 0.760 | 0.698-0.822 | 0.628 | 0.777 | 0.735 | 0.606-0.863 | 0.999 | 0.375 |
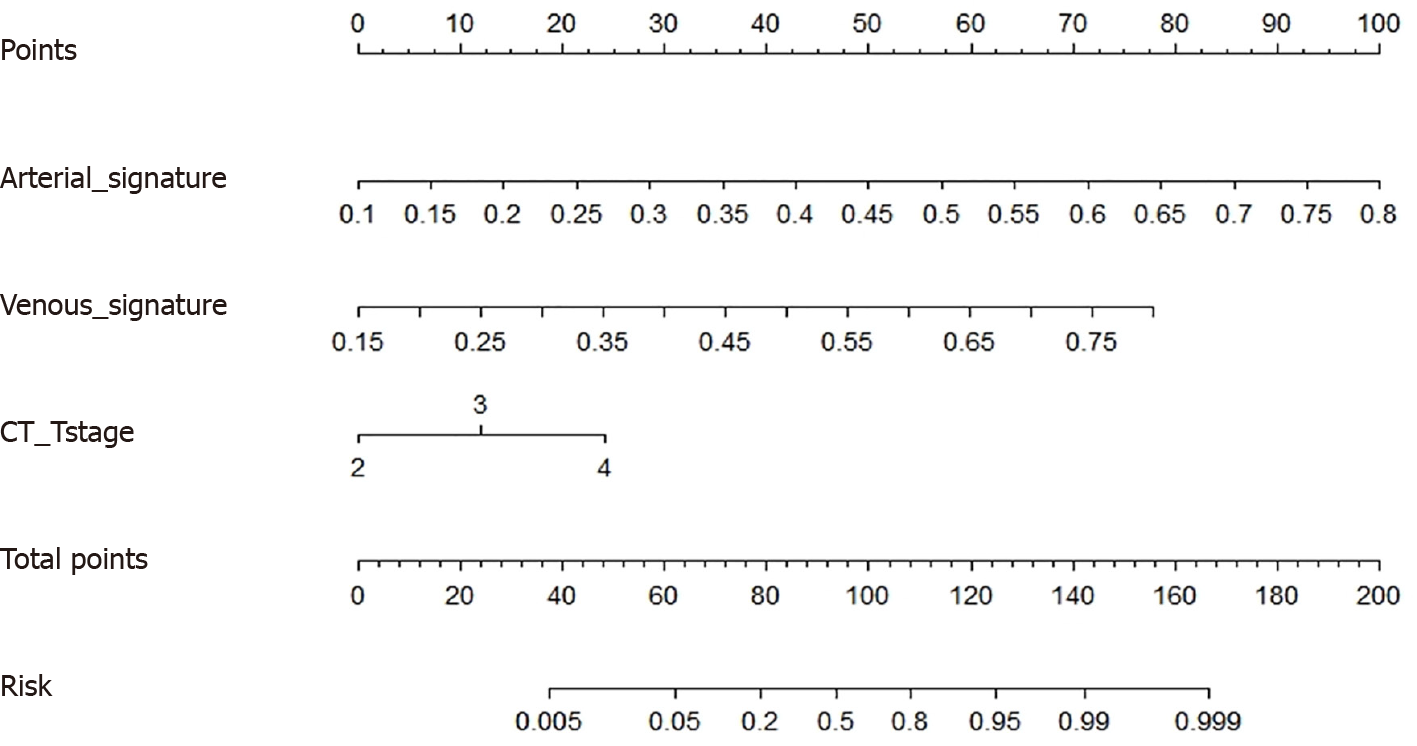
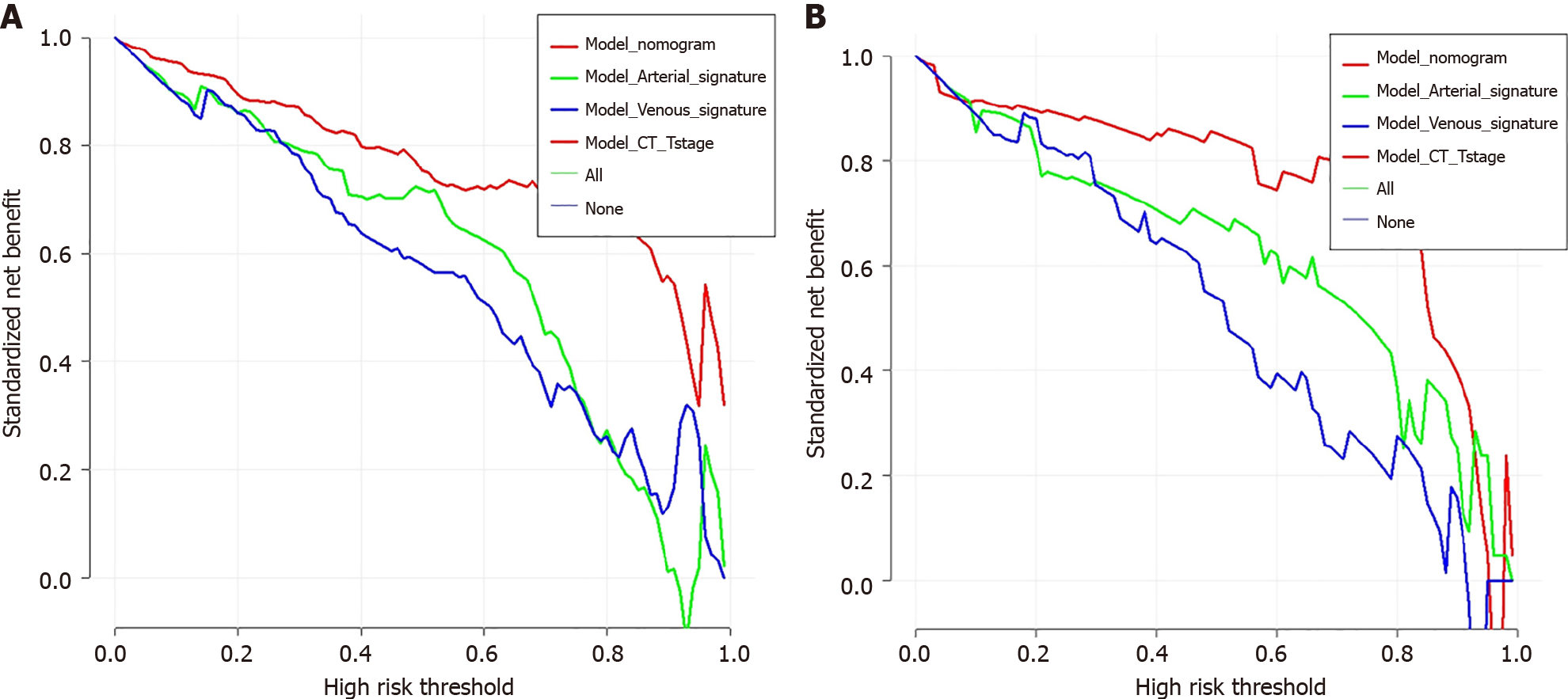
The possibility of the SVM model correctly predicting PNI+ during the venous and arterial phases was noted as Arterial_signature and Venous_signature, respectively. Arterial_signature, Venous_signature, and CT-T stage were merged, and logistic regression was used to create the stacking nomogram (Figure 4). With an AUC of 0.964 (95%CI: 0.944-0.983) in the training group and 0.955 (0.900-0.999) in the test group, the stacking nomogram demonstrated a satisfactory evaluation. In the training group, there were significant differences between the ROC curves of the stacking nomogram and the machine learning models in the arterial and venous phases (P < 0.001); however, there was no significant difference between the stacking nomogram and the ASVM in the test cohort (P = 0.05137). Figure 5 and Tables 3 and 4 show evaluation and comparison of the stacking nomogram, ASVM, VSVM, and CT-T stage. Hosmer–Lemeshow test showed that the stacking nomogram had a good fit (training group: P = 0.867, test group: P = 0.256). The calibration curve of the stacking nomogram is shown in Figure 5, which illustrates that there is good agreement between the predicted and actual values. A strong net benefit is displayed by the decision curve. Figure 6 shows the radiomics model, clinical model, and stacking nomogram decision curve analysis. The decision curve indicated that, if the threshold likelihood of PNI was between 10% and 90%, the stacking nomogram gained more from treating all patient alternatives or from having no treatment options.
| Model | Dataset | AUC | 95%CI | Sensitivity | Specificity | Recall | Accuracy | Precision | F1-score |
| ASVM | Train | 0.904 | 0.865-0.943 | 0.840 | 0.885 | 0.840 | 0.863 | 0.880 | 0.860 |
| Test | 0.890 | 0.794-0.987 | 0.857 | 0.850 | 0.857 | 0.854 | 0.851 | 0.854 | |
| VSVM | Train | 0.890 | 0.850-0.930 | 0.926 | 0.703 | 0.926 | 0.815 | 0.757 | 0.786 |
| Test | 0.867 | 0.778-0.956 | 0.810 | 0.800 | 0.810 | 0.805 | 0.802 | 0.806 | |
| CT-Tstage | Train | 0.647 | 0.583-0.710 | 0.553 | 0.689 | 0.553 | 0.621 | 0.640 | 0.593 |
| Test | 0.730 | 0.607-0.854 | 0.525 | 0.825 | 0.525 | 0.675 | 0.750 | 0.618 | |
| Nomogram | Train | 0.964 | 0.944-0.983 | 0.800 | 0.789 | 0.800 | 0.795 | 0.791 | 0.795 |
| Test | 0.955 | 0.900-0.999 | 0.952 | 0.900 | 0.952 | 0.928 | 0.905 | 0.919 |
| Model | Training set | Test set | ||||||
| Delong-test | ASVM | VSVM | CT-T stage | Nomogram | ASVM | VSVM | CT-T stage | Nomogram |
| ASVM | - | 0.6304 | 1.934e-11 | 0.000271 | - | 0.6997 | 0.0527 | 0.05137 |
| VSVM | - | - | 1.737e-10 | 2.15e-05 | - | - | 0.0692 | 0.03611 |
| CT-Tstage | - | - | - | 2.2e-16 | - | - | - | 0.000305 |
| Nomogram | - | - | - | - | - | - | - | - |
According to recent research, PNI is the result of interactions between tumor and nerve cells as well as different biological signaling chemicals and their receptors inside the peripheral milieu. These interactions may cause the cancer to become more aggressive and to spread[13]. Currently, pathological investigation is the sole method available to ascertain PNI status. Preoperative prediction of PNI facilitates the development of individualized treatment. For instance, postoperative chemotherapy and neoadjuvant chemoradiotherapy can help the majority of PNI+ patients, increasing the survival rate[14-16]. Therefore, accurate preoperative prediction of PNI status helps to evaluate the prognosis of RC patients.
The evaluation of noninvasive prognosis in RC patients has always been a difficult issue. Traditional imaging methods, such as CT and MRI, cannot accurately predict the PNI status of RC; however, RC is a temporally and spatially heterogeneous disease. The risk of invasive sampling and potential complications limit its application in tumor progression and real-time monitoring, so radiomics gradually attracted the attention of oncologists and clinicians. There have been several studies using radiomics to assess PNI status in RC. Chen et al[17] proposed a nomogram model for predicting preo
To construct the radiomics model, we screened 15 and 13 radiomics features highly associated with PNI from the arterial and venous stages, respectively. In both arterial and venous stages, the radiomics features GLSZM-small area emphasis showed a negative correlation with PNI of RC. GLSZM is defined as the number of connecting elements with the same gray intensity. Small area emphasis is a measure of regional distribution of small size, with larger values representing smaller areas and better texture, which indicates that PNI-positive tumors have rough texture features, and more detailed features are needed to describe PNI-negative tumors. These reflect the existence of some specific connection between tumor intensity, differences within tumor texture, and PNI. Previous studies also showed that texture features are predictive in many cases, which is consistent with the results of this study[19,20].
A large number of studies on disease classification, differential diagnosis and predictive prognosis have shown that deep learning can better promote radiomics analysis, which is becoming more widely used in the field of medical imaging. The use of deep learning methods to process and analyze medical imaging data has promoted the development of precision and personalized medicine[21,22]. Deep learning features comprised most of the features that we screened (25 of 28), suggesting that deep learning is more predictively significant in the preoperative PNI prediction of RC. We constructed a neural network by transfer learning using the Resnet-50 method to provide machine learning models with strong feature representation capabilities. Transfer learning is a generalized and efficient method of learning that involves applying knowledge from tasks related to general object recognition to challenges specific to a certain domain. Therefore, the main contribution of this study is to use the ImageNet pretrained deep learning CNN architecture as the foundation to establish an automated tool for the detection and diagnosis of PNI in CT images. Its main idea is to use their CNN structure and its learning weights on ImageNet, and use the ResNet50 architecture to accurately extract features from the ROIs. The Resnet-50 algorithm is based on the residual learning mechanism. The algorithm simplifies the learning process, making it possible to train deeper networks, and solves the problem of gradient dispersion and disappearance as the network deepens. Research has demonstrated that a range of tumor-related tasks, such as tumor diagnosis, classification, grade, stage, and prognostic prediction, as well as identification of pathological features, biomarkers, and genetic alterations, may be carried out using CNN algorithms based on Resnet50. Several studies have also demonstrated the clinical utility of this architecture of Resnet-50[23-25].
Selecting the right machine learning model is essential for maintaining the stability and performance of the model. Many researchers[18,20] only used one machine learning technique to create their models. Chen et al[20] discovered that the logistic-regression-based MR radiomics model was effective in predicting PNI in patients with colorectal cancer (AUC = 0.86 in the training group; AUC = 0.85 in the test group). This study investigated the diagnostic performance of four machine learning models built on the KNN, SVM, MLP, and LR machine learning algorithms. With AUC values greater than KNN, MLP, and LR algorithms (training group: AUC = 0.904; validation group: AUC = 0.890), SVM was the best machine learning method. In the realm of machine learning, SVMs are examples of classical classification algorithms. The SVM is known as the most robust classifier and has good generalization ability, suitability for small samples, and high dimensional features. It has been applied extensively to numerous classification and regression issues, yielding positive outcomes[26,27]. To further improve the ability to predict PNI, we constructed a stacking nomogram model using the input variable obtained by the above SVM machine learning algorithm. The AUC of this stacking nomogram model (training group, AUC = 0.964; test group, AUC = 0.955) improved the AUC compared with a single machine learning model and higher than the recently reported results (training group, AUC = 0.88; test group, AUC = 0.80) [17]. Although the increase in AUC of the stacking nomogram model was not significant compared with the machine learning ASVM model (
Our study had some limitations. This was a retrospective study conducted in a single center, hence adequate external data are required to validate the findings. Our sample size was small, and future research and data from other centers are required to confirm the generalizability of our model. These could be useful directions for future investigation.
We provide a stacking nomogram model of radiomics based on contrast-enhanced CT that may predict the PNI status of RC and provide clinicians with additional quantifiable evidence for the formulation of individualized treatment options.
Thanks to the Fourth Affiliated Hospital of China Medical University for technical support of this study.
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11436] [Article Influence: 3812.0] [Reference Citation Analysis (4)] |
| 2. | Liu Q, Ma Z, Cao Q, Zhao H, Guo Y, Liu T, Li J. Perineural invasion-associated biomarkers for tumor development. Biomed Pharmacother. 2022;155:113691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 3. | Fu Y, Zhang X, Ding Z, Zhu N, Song Y, Zhang X, Jing Y, Yu Y, Huang X, Zhang L, Hu Q, Ni Y, Ding L. Worst Pattern of Perineural Invasion Redefines the Spatial Localization of Nerves in Oral Squamous Cell Carcinoma. Front Oncol. 2021;11:766902. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 4. | Kaya T, Dursun A. Can Lymphovascular and Perineural Invasion be Additional Staging Criteria in Colorectal Cancer? J Coll Physicians Surg Pak. 2021;31:657-662. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 5. | Li M, Jin YM, Zhang YC, Zhao YL, Huang CC, Liu SM, Song B. Radiomics for predicting perineural invasion status in rectal cancer. World J Gastroenterol. 2021;27:5610-5621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 6. | Kim S, Huh JW, Lee WY, Yun SH, Kim HC, Cho YB, Park YA, Shin JK. Prognostic Impact of Lymphatic Invasion, Venous Invasion, Perineural Invasion, and Tumor Budding in Rectal Cancer Treated With Neoadjuvant Chemoradiotherapy Followed by Total Mesorectal Excision. Dis Colon Rectum. 2023;66:905-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 7. | Lafata KJ, Wang Y, Konkel B, Yin FF, Bashir MR. Radiomics: a primer on high-throughput image phenotyping. Abdom Radiol (NY). 2022;47:2986-3002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 54] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 8. | Mayerhoefer ME, Materka A, Langs G, Häggström I, Szczypiński P, Gibbs P, Cook G. Introduction to Radiomics. J Nucl Med. 2020;61:488-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 1018] [Article Influence: 203.6] [Reference Citation Analysis (0)] |
| 9. | Bera K, Braman N, Gupta A, Velcheti V, Madabhushi A. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat Rev Clin Oncol. 2022;19:132-146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 501] [Cited by in RCA: 422] [Article Influence: 140.7] [Reference Citation Analysis (1)] |
| 10. | Guo Y, Wang Q, Guo Y, Zhang Y, Fu Y, Zhang H. Preoperative prediction of perineural invasion with multi-modality radiomics in rectal cancer. Sci Rep. 2021;11:9429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 11. | Ma J, Guo D, Miao W, Wang Y, Yan L, Wu F, Zhang C, Zhang R, Zuo P, Yang G, Wang Z. The value of (18)F-FDG PET/CT-based radiomics in predicting perineural invasion and outcome in non-metastatic colorectal cancer. Abdom Radiol (NY). 2022;47:1244-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 12. | Cerminara SE, Cheng P, Kostner L, Huber S, Kunz M, Maul JT, Böhm JS, Dettwiler CF, Geser A, Jakopović C, Stoffel LM, Peter JK, Levesque M, Navarini AA, Maul LV. Diagnostic performance of augmented intelligence with 2D and 3D total body photography and convolutional neural networks in a high-risk population for melanoma under real-world conditions: A new era of skin cancer screening? Eur J Cancer. 2023;190:112954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 13. | Chen SH, Zhang BY, Zhou B, Zhu CZ, Sun LQ, Feng YJ. Perineural invasion of cancer: a complex crosstalk between cells and molecules in the perineural niche. Am J Cancer Res. 2019;9:1-21. [PubMed] |
| 14. | Kim S, Huh JW, Lee WY, Yun SH, Kim HC, Cho YB, Park YA, Shin JK. Lymphovascular invasion, perineural invasion, and tumor budding are prognostic factors for stage I colon cancer recurrence. Int J Colorectal Dis. 2020;35:881-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 15. | Sung SY, Kim SH, Jang HS, Song JH, Jeong S, Jung JH, Lee JH. Pathologic Implications of Radial Resection Margin and Perineural Invasion to Adjuvant Chemotherapy after Preoperative Chemoradiotherapy and Surgery for Rectal Cancer: A Multi-Institutional and Case-Matched Control Study. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 16. | Kim CH, Yeom SS, Lee SY, Kim HR, Kim YJ, Lee KH, Lee JH. Prognostic Impact of Perineural Invasion in Rectal Cancer After Neoadjuvant Chemoradiotherapy. World J Surg. 2019;43:260-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 17. | Chen Q, Cui Y, Xue T, Peng H, Li M, Zhu X, Duan S, Gu H, Feng F. Computed tomography-based radiomics nomogram for the preoperative prediction of perineural invasion in colorectal cancer: a multicentre study. Abdom Radiol (NY). 2022;47:3251-3263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 18. | Yang YS, Qiu YJ, Zheng GH, Gong HP, Ge YQ, Zhang YF, Feng F, Wang YT. High resolution MRI-based radiomic nomogram in predicting perineural invasion in rectal cancer. Cancer Imaging. 2021;21:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 19. | Zhang Y, Peng J, Liu J, Ma Y, Shu Z. Preoperative Prediction of Perineural Invasion Status of Rectal Cancer Based on Radiomics Nomogram of Multiparametric Magnetic Resonance Imaging. Front Oncol. 2022;12:828904. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 20. | Chen J, Chen Y, Zheng D, Pang P, Zhang H, Zheng X, Liao J. Pretreatment MR-based radiomics nomogram as potential imaging biomarker for individualized assessment of perineural invasion status in rectal cancer. Abdom Radiol (NY). 2021;46:847-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 21. | Bo L, Zhang Z, Jiang Z, Yang C, Huang P, Chen T, Wang Y, Yu G, Tan X, Cheng Q, Li D, Liu Z. Differentiation of Brain Abscess From Cystic Glioma Using Conventional MRI Based on Deep Transfer Learning Features and Hand-Crafted Radiomics Features. Front Med (Lausanne). 2021;8:748144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Wang H, Wang L, Lee EH, Zheng J, Zhang W, Halabi S, Liu C, Deng K, Song J, Yeom KW. Decoding COVID-19 pneumonia: comparison of deep learning and radiomics CT image signatures. Eur J Nucl Med Mol Imaging. 2021;48:1478-1486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 23. | Elpeltagy M, Sallam H. Automatic prediction of COVID- 19 from chest images using modified ResNet50. Multimed Tools Appl. 2021;80:26451-26463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 24. | Sharma AK, Nandal A, Dhaka A, Koundal D, Bogatinoska DC, Alyami H. Enhanced Watershed Segmentation Algorithm-Based Modified ResNet50 Model for Brain Tumor Detection. Biomed Res Int. 2022;2022:7348344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 25. | Hossain MB, Iqbal SMHS, Islam MM, Akhtar MN, Sarker IH. Transfer learning with fine-tuned deep CNN ResNet50 model for classifying COVID-19 from chest X-ray images. Inform Med Unlocked. 2022;30:100916. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 26. | Dong D, Fang MJ, Tang L, Shan XH, Gao JB, Giganti F, Wang RP, Chen X, Wang XX, Palumbo D, Fu J, Li WC, Li J, Zhong LZ, De Cobelli F, Ji JF, Liu ZY, Tian J. Deep learning radiomic nomogram can predict the number of lymph node metastasis in locally advanced gastric cancer: an international multicenter study. Ann Oncol. 2020;31:912-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 269] [Article Influence: 53.8] [Reference Citation Analysis (1)] |
| 27. | Aswathy MA, Jagannath M. An SVM approach towards breast cancer classification from H&E-stained histopathology images based on integrated features. Med Biol Eng Comput. 2021;59:1773-1783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |