Published online Apr 28, 2022. doi: 10.35711/aimi.v3.i2.21
Peer-review started: December 16, 2021
First decision: January 26, 2022
Revised: February 12, 2022
Accepted: March 16, 2022
Article in press: March 16, 2022
Published online: April 28, 2022
Processing time: 133 Days and 0.6 Hours
Pancreatic adenocarcinoma remains to be one of the deadliest malignancies in the world despite treatment advancement over the past few decades. Its low survival rates and poor prognosis can be attributed to ambiguity in recommendations for screening and late symptom onset, contributing to its late presentation. In the recent years, artificial intelligence (AI) as emerged as a field to aid in the process of clinical decision making. Considerable efforts have been made in the realm of AI to screen for and predict future development of pancreatic ductal adenocarcinoma. This review discusses the use of AI in early detection and screening for pancreatic adenocarcinoma, and factors which may limit its use in a clinical setting.
Core Tip: Pancreatic adenocarcinoma has poor survival rate and high morbidity. Artificial intelligence is a potential tool to screen for high risk individuals and for early detection of pancreatic adenocarcinoma. Despite advances made in artificial intelligence research in pancreatic adenocarcinoma, it faces a number of challenges before it can be generalised and applied in a clinical setting.
- Citation: Lin KW, Ang TL, Li JW. Role of artificial intelligence in early detection and screening for pancreatic adenocarcinoma. Artif Intell Med Imaging 2022; 3(2): 21-32
- URL: https://www.wjgnet.com/2644-3260/full/v3/i2/21.htm
- DOI: https://dx.doi.org/10.35711/aimi.v3.i2.21
The global incidence of pancreatic cancer is increasing, and it remains as one of the leading causes of cancer-related death, with 495773 new cases of pancreatic cancer diagnosed and accounting for 466003 deaths in 2020[1]. Although the 5-year survival rates for pancreatic ductal adenocarcinoma (PDAC) have improved, it remains low at approximately 9%[2,3], and the overall prognosis of PDAC is poor. This is partly due to the late stage of presentation of PDAC, which is largely dependent on patient symptoms for suspicion of the disease[4,5]. Early cases are asymptomatic and there is a lack of a simple screening tool for clinical use unlike the case of colorectal cancer screening where screening can be performed in the primary care setting with the use of fecal immunohistochemical test. In the case of PDAC, cross-sectional imaging tests such as computed tomography (CT) or magnetic resonance imaging (MRI) are needed for detection, making widespread population screening unfeasible. Germline mutations and a family history of PDAC have been identified as the strongest risk factors for the disease[6,7]. As such, efforts in screening programmes have focused their attention on this group of patients[8]. Pancreatic cysts, increased age, and smoking are also known risk factors for PDAC[5,9,10], although it may not be practical to conduct routine surveillance for patients with these risk factors. There is an interest in selecting higher risk patients for screening, as the appropriate use biomarkers and imaging may result in detection of early-stage PDAC amenable to curative resection[2,3,11-15].
Artificial intelligence (AI) is a branch in computer science where computer systems are designed to perform tasks which would require human intelligence. It is recognised as a potential tool as part of the screening efforts and building predictive models[16]. Most progress for AI in endoscopy has been made in the field of colonoscopy, where polyp detection and characterisation has been studied[17]. Computer-aided diagnosis has also been extended to detection and screening of PDAC[18] in endoscopic ultrasound (EUS)[19,20], MRI[21] and cytology from fine needle sampling[22]. In recent years, various groups have harnessed the potential of AI in creating prediction models. These include The Felix Project[23], the Pancreatic-Cancer Collective[24], and the Early Detection Research Network[25] effort.
This mini-review aims to study the role of AI in the early detection and screening for pancreatic cancer, as well as factors which may limit its use.
A comprehensive literature search was performed in the PubMed, MEDLINE and EMBASE electronic databases from the inception of the databases up to and including 30 November 2021. The key words used were “artificial intelligence”, “pancreatic cancer”, “pancreatic adenocarcinoma”, “pancreatic ductal adenocarcinoma”, “pancreatic carcinoma”, “screening”, and “early detection”. These were supplemented with manual searches of references from retrieved articles. Publications in English were considered for this mini-review.
AI is a term that refers to the ability of a computer programme to imitate the human mind to perform tasks such as problem solving and learning[26,27].
Machine learning (ML) is the commonest branch of AI used in medicine and refers to a mathematical model that aims to generate a prediction based on a set of data provided[28,29]. In supervised learning, the data points are labelled and the ML model “learns” from these labels and identifies new data points. In contrast, labels are not provided in unsupervised learning, and the model recognises the patterns of the data by learning its unknown properties and identifying crucial data checkpoints. This is especially important when the gold standard is not available[29].
Deep learning (DL) is subset of ML that employs the use of Artificial Neural Networks (ANN). Like the human brain, ANN consists of layers of artificial neurons that are interlinked. Each layer receives a weighted signal from the previous layer(s) and these signals will be propagated to the next layer when a specific threshold is exceeded[29]. In the setting of a pancreatic lesion or cancer, DL first identifies the basics of the lesion (e.g., location) in its initial layers before moving on to next layer for further characterisation (e.g., size, shape, colour). A final prediction of the pancreatic lesion is made after a systematic assessment via multiple layers of neural network[29].
ANNs are first trained using the training data set, where the model learns to identify specific patterns to obtain a relationship between the input and the output. Hyperparameters refer to all settings that are pre-determined by the investigator and are used to construct the model for optimal execution of a particular task or on a specific dataset. The validation data set involves a different data set that is used to fine-tune the hyperparameters of the model. Finally, the test data set refers to a data set whose purpose is to evaluate the performance of the model against unseen data and determine its generalizability[29]. This set needs to be unseen by the model during training and validation. However in certain studies, the test set is sometimes a subset of the training or validation data set, which many result in overfitting of the model. This may lead to a discrepancy in the performance of the model when tested in the same centre and a decline in performance when validated externally.
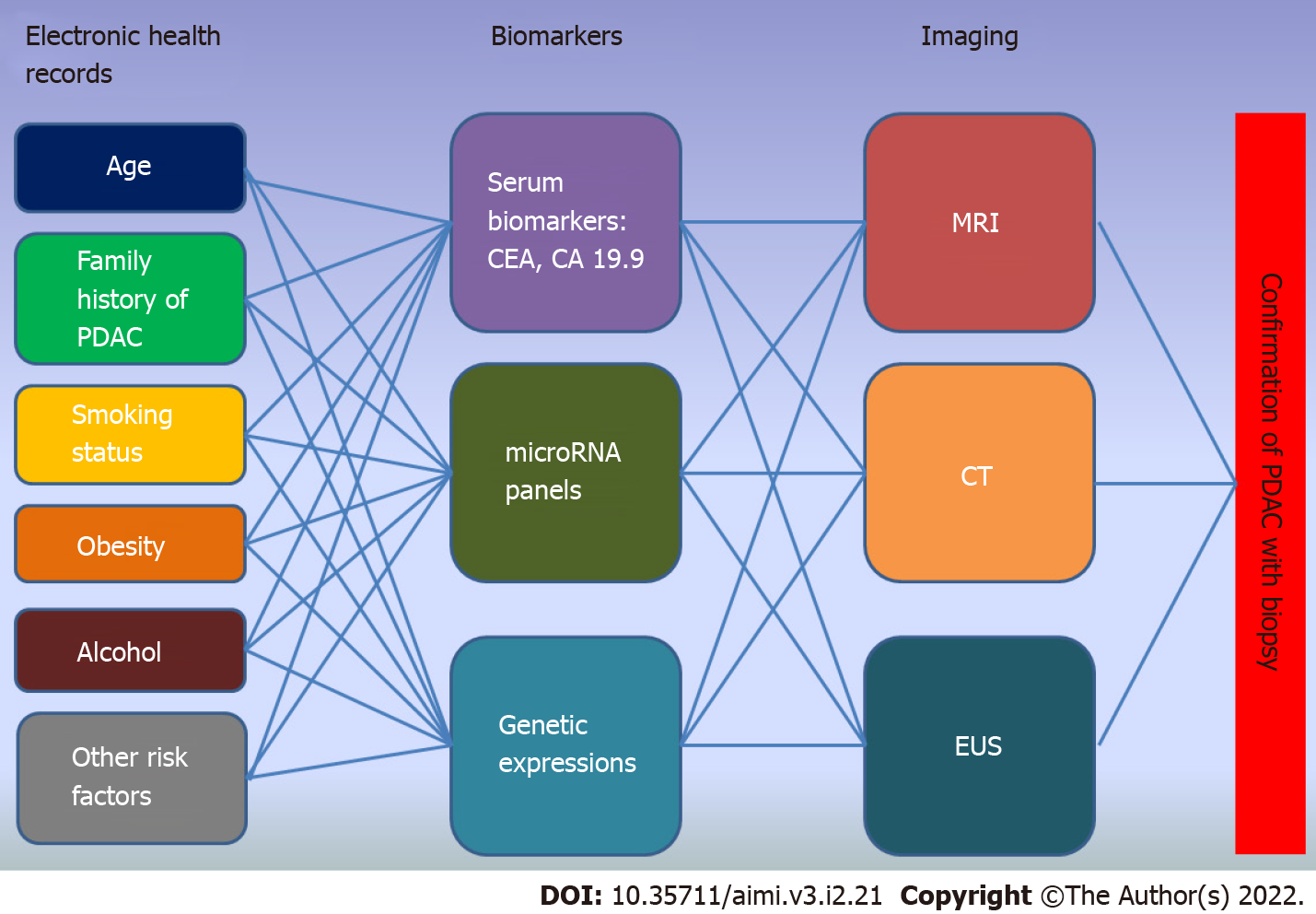
Early detection of pancreatic cancer requires a step wise approach in order to systematically screen for risk factors and identify high-risk groups. Figure 1 is a schematic diagram showing the workflow and neural network to be designed for an early detection protocol. It represents the complex interplay between each of the input(s) to be processed for the next neural layer(s) until a final output is obtained. We will be discussing the role of AI in early detection of pancreatic cancer based on this model.
The identification of risk factors for pancreatic cancer is essential in identifying the specific population which would benefit from screening[18,30,31]. Factors such as diabetes, hemoglobin A1C (HbA1c) value, weight, body mass index (BMI), blood type, smoking status, alcohol use and family history of pancreatic cancer influence the age of onset of screening for an individual[13,32]. These factors are easily available in the primary care setting and could potentially predict the development of pancreatic cancer within 5 years, even before any changes to the pancreas can be detected on imaging[30]. However, most of the data is stored in health records, which are often proprietary or internet-separated to protect patient data. The retrieval and subsequent integration of data from different platforms remains a manual and laborious process for physicians[30]. Even after retrieval, there are no validated scoring systems to assess these risk factors and stratify patients. On the other hand, AI, with the aid of Natural Language Processing, can facilitate this process[33-38]. In a case-control study, Malhotra et al[33] created an algorithm based on electronic health records (EHR) obtained from primary care to identify 41.3% of patients (≤ 60 years old) who had significant risk of developing pancreatic cancer up to 20 mo prior to diagnosis with a sensitivity, specificity, area under the receiver operating characteristic (AUROC) curve of 72.5%, 59.0% and 0.66%, respectively. Similarly, Appelbaum et al[35] was able to train an ANN using 101381 EHRs to predict the development of PDAC one year before the diagnosis in a population of high-risk patients (AUROC 0.68, confidence interval (CI): 0.65-0.71).
Despite its potential benefits, research in AI for the above purpose is still preliminary as they are mostly based on retrospective data from single institutions or registries, and hence not ready for use in a wider clinical setting[33-38]. One of the major limitations would be the lack validation in the real-world setting or at least in populations derived from different centres to overcome the risk of bias and overfitting.
The use of AI in EHR faces other challenges. Various institutions’ medical records are built on different healthcare systems and encoding systems, making the task of harmonising them difficult[30]. There is also a lack of standardised clinical research data collection models. To overcome this, efforts are made to build a model of processing and integrating data across institutions. The i2b2 was created to review medical records, retrieve specific data of interest and repurpose it for research[39]. The Observational Health Data Sciences and Informatics was developed from the Observational Medical Outcomes Partnership, an initiative that develops the Common Data Model aiming to gather information from different data sets or medical repositories and systemically analyse them in a common platform[40]. Similarly, the National Patient-centered Clinical research network is another example which was developed in United States to access millions of EHR and create a common data set for research purposes[41]. A common dataset with a standardised format for input of data relevant to PDAC would enable AI systems to leverage on big data to identify changing risk profiles in PDAC, enabling the clinician to channel resources for screening to the appropriate cohorts of patients depending on the population from which this data has been derived.
While these are upcoming and promising initiatives, concerns surrounding restrictions in data sharing, privacy issues, and maintenance costs could hinder data collection efforts[18]. EHRs are also stored in different languages in different regions of the world, making the integration of data difficult. Besides, once data sets are gathered, obtaining IRB approval from the various sites for research may be difficult.
Carbohydrate Antigen 19-9 (CA19-9) and carcinoembryonic antigen (CEA) are the most widely used markers for screening of PDAC, but have also been proven to lack the specificity when applied individually and without clinical context[42,43]. On the other hand, a combined measurement can potentially increase its sensitivity and specificity up to 1 year before the diagnosis of PDAC[44-46]. Capitalising on this concept, Yang et al[47], developed an algorithm (with 658 subjects in its training set) to diagnose pancreatic cancer by using ANN to combine CA19-9, CA125 and CEA values. This model was subsequently evaluated against the test set and was able to yield an AUROC of 0.905 (95%CI = 0.868-0.942) and a high diagnostic accuracy of 83.5% for pancreatic cancer.
New biomarkers for PDAC such as MicroRNAs and gene expressions have generated much interest in the recent years[45,48-52]. MicroRNAs are non-coding RNAs that are involved in the regulation of biological pathways, and when altered, could lead to the development of PDAC[53]. MicroRNAs can potentially predict future PDAC[54] or detect early stage pancreatic cancer. However, they have the same limitations in sensitivity and specificity when applied without clinical context and as independent test[55,56]. A combination of the commonly used biomarkers and newer biomarkers may address the problem of low sensitivity and specificity[56], and in particular can be combined with clinical and demographic information as described earlier to increase its usefulness.
While AI is able to make use of plasma microRNA panels and specific gene expressions to diagnose pancreatic cancer[57,58], studies on their use on predicting future pancreatic cancer are not available[55]. By integrating Particle Swarm Optimization, ANN and Neighborhood Component Analysis iterations on a list of microRNAs that are most commonly expressed by pancreatic cancer, Alizadeh et al[59] created a model consisting of 5 MicroRNAs (miR-663a, miR-1469, miR-92a-2-5p, miR-125b-1-3p and miR-532-5p) to diagnose pancreatic cancer (Accuracy: 0.93, Sensitivity: 93%, and Specificity: 92%). Similarly in a multicentre study by Cao et al[57], a machine learning approach was able to identify 2 panels of microRNAs to differentiate pancreatic cancer from chronic pancreatitis with an accuracy of above 80%.
Gene expressions have gained popularity in diagnosing pancreatic cancer[13,60]. Using a machine learning approach, Khatri et al[61] analysed the results from transcriptomics-based meta-analysis to create a nine-gene panel to diagnose pancreatic cancer. This panel was able to differentiate PDAC from chronic pancreatitis with a specificity of 89%, sensitivity of 78%, and accuracy of 83% and an AUROC of 0.95. As compared to a normal pancreas, it was also used to identify stage I and II PDACs with a sensitivity of 74%, specificity of 75%, and an AUROC of 0.82. In another study, a machine learning algorithm was formulated based on the biochemical differences in the serum of 2 groups of subjects (PDAC group and High risk group) detected via the use of Probe Electrospray Ionization Mass Spectrometry (PESI-MS) to identify early stages of pancreatic cancer[62]. It was able to differentiate healthy controls from subjects with earlier stage of PDAC with sensitivity of 81.2% and specificity of 96.8% respectively and an accuracy of 92.9%.
At present, these studies have shown that AI can offer the advantage of identifying specific microRNA and genetic combinations to identifying pancreatic cancer at a faster speed, making this process less laborious. However, these studies lack external validation, limiting their application in modern practice. Besides, studies utilising AI to formulate specific sequences to accurately predict future pancreatic cancer development are still lacking. More studies are required to analyse its ability in predicting future pancreatic cancer for high risk groups especially during the latency period.
Various studies have been conducted using AI to diagnose pancreatic cancer and yielded promising results. Table 1 summarises the studies to date[21,63-75]. In a retrospective study, Liu et al[69] was able to train a convolutional neural network (CNN) to identify pancreatic cancer on contrast-enhanced CT and achieve an AUROC of 0.9, with more than 90% for its sensitivity and specificity for its test set. It maintained good sensitivity of 91.3%, specificity of 84.5%, an accuracy of 85.6% and AUROC of 0.955 (95%CI 0.955-0.956) with the validation set. Further analysis revealed that with CNN, radiologists missed 7% of the pancreatic cancers, of which majority were accurately diagnosed by CNN[69]. By enhancing the CNN, Liu et al[73] was able to process the CT images and obtain the diagnosis faster than the radiologists (3 s for CNN vs 8 mins for a radiologist) with an AUROC of 0.9632, proving that AI is comparable to radiologists.
| Ref. | Clinical question | Training set (number of subjects) | Validation set (number of subjects) | AI instrument | AUROC | Accuracy | Sensitivity | Specificity |
| Watson et al[66], 2021 | Detection of pancreatic cystic neoplasms (including PDAC) vs benign cysts | 18 | 9 | CNN | NA | NA | NA | NA |
| Si et al[65], 2021 | Detection of pancreatic cancer (including PDAC, IPMN, PNET) | 319 | 347 | DL | 0.871 | 87.6% for PDAC | 86.8% for pancreatic cancer | 69.5% for pancreatic cancer |
| Park et al[64], 2020 | Distinguishing pancreatic cancer tissue from autoimmune pancreatitis | 120 | 62 | Random forest machine learning | 0.975 | 95.2% | 89.7% | 100% |
| Ma et al[63], 2020 | Differentiate pancreatic cancer from benign tissue | 330 | 41 | CNN | 0.9653 (plain scan) | 95.47% (plain scan),95.76% (arterial scan), 95.15% (venous phase) | 91.58% (plain scan), 94.08% (arterial scan), 92.28% (venous phase) | 98.3% (plain scan), 97.6% (arterial scan), 97.9% (venous phase) |
| Zhang et al[67], 2020 | Detection of pancreatic cancer | 2650 images | 240 images | CNN | 0.9455 | 90.2% | 83.8% | 91.8% |
| Liu et al[69], 2020 | Differentiating pancreatic cancer tissue from non-cancerous pancreatic tissue | 412 | 139 | CNN | 0.92 | 83.2% | 79.0% | 97.6% |
| Gao et al[71], 2020 | To differentiate pancreatic diseases in pancreatic lesions | 398 | 106 | CNN | 0.9035 (includes PDAC, adenosquamous carcinoma, acinar cell carcinoma, colloid carcinoma, myoepithelial carcinoma, undifferentiated carcinoma with osteoclast-like giant cells, mucinous cystadenocarcinoma, pancreatoblastoma, pancreatic neuroendocrine carcinoma and metastatic carcinoma) | NA | NA | NA |
| Chu et al[70], 2019 | Differentiating PDAC from normal pancreas | 255 | 125 | Random forest | NA | 93.6% | 95% | 92.3% |
| Zhu et al[72], 2019 | Detecting PDAC from normal pancreas | 205 | 234 | CNN | NA | 57.3% | 94.1% | 98.5% |
| Liu et al[73], 2019 | Diagnosis of pancreatic cancer | 238 | 100 | CNN | 0.9632 | NA | NA | NA |
| Corral et al[21], 2019 | Identify and stratify IPMN lesions | 139 | DL | 0.783 | NA | 75% (for PDAC or high grade dysplasia) | 78% (for PDAC or high grade dysplasia) | |
| Chu et al[74], 2019 | Differentiating PDAC from normal pancreas | 456 | DL | NA | NA | 94.1% | 98.5% | |
| Fu et al[75], 2018 | Pancreas segmentation (including PDAC, IPMN, Pancreatic Neuroendocrine Tumors, Serous Cyst Adenoma, and Solid Pseudopapillary Tumour of the pancreas) | 59 | CNN | NA | NA | 82.5% | 76.22 (PPV) | |
Besides CT, EUS has been frequently utilised to diagnosed pancreatic cancer. Table 2 summaries these studies[19,20,76-86]. The EUS-CAD based CNN was developed in a retrospective study by Tonozuka et al[83] to identify lesions harbouring pancreatic cancer in patients with chronic pancreatitis with a sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of 90.2%, 74.9%, 80.1%, and 88.7%, respectively, and an AUROC of 0.924. Similar findings were also echoed in Zhu et al[86] who utilised SVM to obtain a sensitivity, specificity, PPV and NPV of over 90% for diagnosis of pancreatic cancer in chronic pancreatitis.
| Ref. | Clinical question | Training set (number of subjects) | Validation set (number of subjects) | AI instrument | AUROC | Accuracy | Sensitivity | Specificity | |
| Udristoiu et al[84], 2021 | Detecting focal pancreatic masses in four EUS imaging modalities | 65 | CNN and Long Short-term Memory models | 0.97 | 97.6% | 98.1% | 96.7% | ||
| Tonozuka et al[83], 2021 | Detecting PDAC in patients with normal pancreas/Chronic pancreatitis | 92 | CNN | 0.924 | NA | 90.2% | 74.9% | ||
| Marya et al[78], 2021 | Differentiate AIP from PDAC, chronic pancreatitis and other pancreatic diseases | 336 | 124 | CNN | 0.976 | NA | 95% | 90% | |
| Kuwahara et al[77], 2019 | Predicting malignancy in IPMN | 50 | CNN | 0.98 | 94% | 95.7% | 92.6% | ||
| Ozkan et al[80], 2016 | Differentiating pancreatic cancer from healthy pancreas | 260 images | 72 images | ANN | NA | 87.5% | 83.3% | 93.3% | |
| Saftoiu et al[81], 2015 | Differentiate pancreatic cancer from chronic pancreatitis | 117 | 25 | ANN | NA | NA | 94.6% | 94.4% | |
| Zhu et al[86], 2013 | Differentiating pancreatic cancer from chronic pancreatitis. | 194 | 194 | SVM | NA | 94.2% | 96.3% | 93.4% | |
| Saftoiu et al[82], 2012 | Diagnosis of focal pancreatic lesions | 258 patients | ANN | 0.94 | 84.27% | 87.59% | 82.94% | ||
| Zhang et al[85], 2010 | Differentiate pancreatic cancer from non-tumorous tissue | 108 | 108 | SVM | NA | 97.98% | 94.3% | 99.45% | |
| Saftoiu et al[20], 2008 cancer | Differentiate normal pancreas, chronic pancreatitis, pancreatic cancer, and neuroendocrine tumors | 68 | Neural network | 0.847 (for PDAC vs chronic pan-creatitis) | 86.1% (for PDAC vs chronic pan-creatitis) | 93.8% (for PDAC vs chronic pan-creatitis) | 63.6% (for PDAC vs chronic pan-creatitis) | ||
| Das et al[19], 2008 | Differentiating pancreatic adenocarcinoma from non-neoplastic tissue (includes normal pancreas and chronic pancreatitis) | 160 | 159 | ANN | 0.93 | NA | 93% | 92% | |
| Norton et al[79], 2001 | Differentiate malignancy from pancreatitis | 35 | ML | NA | 80% | 100% | 50% | ||
Despite numerous studies looking at using AI to diagnose pancreatic cancer (as shown in Tables 1 and 2), only a few attempted to predict the development to pancreatic cancer. On average, CT changes for early pancreatic cancer starts approximately 12 to 18 mo before diagnosis[87]. Yet, pancreatic cancer can advance from being undetectable to metastatic in a short period of time even before the next surveillance imaging[88,89]. AI-based imaging itself cannot be used to predict pancreatic cancer and should be combined with other markers.
An ideal AI model for predicting pancreatic cancer is one that integrates multiple biochemical, radiological and clinical data[90]. In a retrospective proof-of-concept study, Springer et al[91] developed a supervised machine learning-based approach (CompCyst) based on a combination of patient-reported symptoms, imaging results (including CT, MRI and EUS images), cyst fluid and molecular characteristics to calculate its malignant potential and subsequently determine the management of pancreatic cyst(s). When tested against the validation set, CompCyst outperformed the current standard of care (accuracy 56%) in its ability to identify patients who required surgery, close monitoring or can be discharged (accuracy 69%). CompCyst correctly identified 60% of the surgeries that were not warranted and could have been avoided, while not compromising on its ability to identifying those who truly require surgery. With CompCyst, 71% of the pancreatic lesions were correctly identified as PDAC as compared to 58% based on clinical suspicion[91].
While this study has proven that AI has the potential to incorporate various clinical characteristics, biomarkers, and imaging characteristics to assess for the malignant potential of a pancreatic lesion, it has a number of limitations. Firstly, the imaging characteristics and molecular biomarkers that were identified as high risk features were obtained at the time of surgery and not during screening. These features may not be present early enough to be identified by routine screening. Secondly, important risk factors (including age and diabetes) that were crucial in the early detection of PDAC (as shown in Figure 1) were not included in its learning process, representing a missed step in the screening process. Finally, CompCyst is yet to be externally validated and cannot be applied to the clinical setting currently.
While CompCyst is a potential tool to aid in clinical decision making, future studies aiming at early detection of PDAC face a myriad of challenges. Firstly, the pancreas is a complex organ. Unlike the other organs, the pancreas can be highly variable in its anatomy and location. Moreover, the training data set is highly dependent on the quality of the images provided. Hence, automated segmentation of the pancreas via a deep learning approach remains challenging[92]. Secondly, the lack of databases limits the ability to develop new training sets. There are currently only a few open-access databases[93], and there are issues regarding sharing of images across various institutions as pointed out by the Alliance of Pancreatic Cancer Consortia imaging working group[90]. Finally, the algorithm for early detection of PDAC will have to evaluate images of pancreatic lesion(s) across different time points of surveillance and from different 3 imaging modalities (namely CT, MRI, and EUS). Unlike CompCyst which looks at images at one time point (i.e. at surgery), combining multiple images obtained from periodical surveillance via these 3 imaging modalities will require a very large database and multiple layers.
There is a major gap that needs to be bridged before AI systems for early detection of pancreatic cancer can be developed. Given sufficient training data and cooperation, AI-based image analyzers could match or even outperform physicians in image classification and lesion detection[90].
Despite the recent advances to predict future PDAC, the use of AI in screening for pancreatic cancer remains limited in the clinical setting. Much of the efforts are made in the research setting and lack external validation and generalisability. However, this field remains promising as we recognise the challenges ahead to bridge the necessary gaps. The hope to develop an integrated AI model to screen for PDAC remains a reality, and it will play a complementary role in assisting physicians in their clinical decision making process but not replace it.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Singapore
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Nazari N, Iran; Zhang JX, China S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
| 1. | GLOBOCAN. International Agency for Research on Cancer. 2020. |
| 2. | Blackford AL, Canto MI, Klein AP, Hruban RH, Goggins M. Recent Trends in the Incidence and Survival of Stage 1A Pancreatic Cancer: A Surveillance, Epidemiology, and End Results Analysis. J Natl Cancer Inst. 2020;112:1162-1169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 149] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 3. | Huang L, Jansen L, Balavarca Y, Babaei M, van der Geest L, Lemmens V, Van Eycken L, De Schutter H, Johannesen TB, Primic-Žakelj M, Zadnik V, Besselink MG, Schrotz-King P, Brenner H. Stratified survival of resected and overall pancreatic cancer patients in Europe and the USA in the early twenty-first century: a large, international population-based study. BMC Med. 2018;16:125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 101] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 4. | Gobbi PG, Bergonzi M, Comelli M, Villano L, Pozzoli D, Vanoli A, Dionigi P. The prognostic role of time to diagnosis and presenting symptoms in patients with pancreatic cancer. Cancer Epidemiol. 2013;37:186-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 5. | Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039-1049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1500] [Cited by in RCA: 1703] [Article Influence: 154.8] [Reference Citation Analysis (0)] |
| 6. | Abe T, Blackford AL, Tamura K, Ford M, McCormick P, Chuidian M, Almario JA, Borges M, Lennon AM, Shin EJ, Klein AP, Hruban RH, Canto MI, Goggins M. Deleterious Germline Mutations Are a Risk Factor for Neoplastic Progression Among High-Risk Individuals Undergoing Pancreatic Surveillance. J Clin Oncol. 2019;37:1070-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 7. | Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol. 2021;18:493-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 674] [Article Influence: 168.5] [Reference Citation Analysis (0)] |
| 8. | Capurso G, Paiella S, Carrara S, Butturini G, Secchettin E, Frulloni L, Zerbi A, Falconi M. Italian registry of families at risk of pancreatic cancer: AISP Familial Pancreatic Cancer Study Group. Dig Liver Dis. 2020;52:1126-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Crippa S, Bassi C, Salvia R, Malleo G, Marchegiani G, Rebours V, Levy P, Partelli S, Suleiman SL, Banks PA, Ahmed N, Chari ST, Fernández-Del Castillo C, Falconi M. Low progression of intraductal papillary mucinous neoplasms with worrisome features and high-risk stigmata undergoing non-operative management: a mid-term follow-up analysis. Gut. 2017;66:495-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 151] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 10. | Weissman S, Takakura K, Eibl G, Pandol SJ, Saruta M. The Diverse Involvement of Cigarette Smoking in Pancreatic Cancer Development and Prognosis. Pancreas. 2020;49:612-620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 11. | Canto MI, Almario JA, Schulick RD, Yeo CJ, Klein A, Blackford A, Shin EJ, Sanyal A, Yenokyan G, Lennon AM, Kamel IR, Fishman EK, Wolfgang C, Weiss M, Hruban RH, Goggins M. Risk of Neoplastic Progression in Individuals at High Risk for Pancreatic Cancer Undergoing Long-term Surveillance. Gastroenterology. 2018;155:740-751.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 303] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 12. | Aslanian HR, Lee JH, Canto MI. AGA Clinical Practice Update on Pancreas Cancer Screening in High-Risk Individuals: Expert Review. Gastroenterology. 2020;159:358-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 179] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 13. | Gonda TA, Everett JN, Wallace M, Simeone DM; PRECEDE Consortium. Recommendations for a More Organized and Effective Approach to the Early Detection of Pancreatic Cancer From the PRECEDE (Pancreatic Cancer Early Detection) Consortium. Gastroenterology. 2021;161:1751-1757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 14. | Kenner BJ, Chari ST, Maitra A, Srivastava S, Cleeter DF, Go VL, Rothschild LJ, Goldberg AE. Early Detection of Pancreatic Cancer-a Defined Future Using Lessons From Other Cancers: A White Paper. Pancreas. 2016;45:1073-1079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 15. | Overbeek KA, Levink IJM, Koopmann BDM, Harinck F, Konings ICAW, Ausems MGEM, Wagner A, Fockens P, van Eijck CH, Groot Koerkamp B, Busch ORC, Besselink MG, Bastiaansen BAJ, van Driel LMJW, Erler NS, Vleggaar FP, Poley JW, Cahen DL, van Hooft JE, Bruno MJ; Dutch Familial Pancreatic Cancer Surveillance Study Group. Long-term yield of pancreatic cancer surveillance in high-risk individuals. Gut. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Richter AN, Khoshgoftaar TM. A review of statistical and machine learning methods for modeling cancer risk using structured clinical data. Artif Intell Med. 2018;90:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 17. | Li JW, Ang TL. Colonoscopy and artificial intelligence: Bridging the gap or a gap needing to be bridged? AIGE. 2021;2:36-49. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Kenner B, Chari ST, Kelsen D, Klimstra DS, Pandol SJ, Rosenthal M, Rustgi AK, Taylor JA, Yala A, Abul-Husn N, Andersen DK, Bernstein D, Brunak S, Canto MI, Eldar YC, Fishman EK, Fleshman J, Go VLW, Holt JM, Field B, Goldberg A, Hoos W, Iacobuzio-Donahue C, Li D, Lidgard G, Maitra A, Matrisian LM, Poblete S, Rothschild L, Sander C, Schwartz LH, Shalit U, Srivastava S, Wolpin B. Artificial Intelligence and Early Detection of Pancreatic Cancer: 2020 Summative Review. Pancreas. 2021;50:251-279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 19. | Das A, Nguyen CC, Li F, Li B. Digital image analysis of EUS images accurately differentiates pancreatic cancer from chronic pancreatitis and normal tissue. Gastrointest Endosc. 2008;67:861-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 81] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 20. | Săftoiu A, Vilmann P, Gorunescu F, Gheonea DI, Gorunescu M, Ciurea T, Popescu GL, Iordache A, Hassan H, Iordache S. Neural network analysis of dynamic sequences of EUS elastography used for the differential diagnosis of chronic pancreatitis and pancreatic cancer. Gastrointest Endosc. 2008;68:1086-1094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 173] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 21. | Corral JE, Hussein S, Kandel P, Bolan CW, Bagci U, Wallace MB. Deep Learning to Classify Intraductal Papillary Mucinous Neoplasms Using Magnetic Resonance Imaging. Pancreas. 2019;48:805-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 22. | Momeni-Boroujeni A, Yousefi E, Somma J. Computer-assisted cytologic diagnosis in pancreatic FNA: An application of neural networks to image analysis. Cancer Cytopathol. 2017;125:926-933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | The Lustgarten Foundation. Deep Learning for Radiologists: A Beginner's Guide 2021. Available from: https://www.ctisus.com/responsive/deep-learning/felix.asp. |
| 24. | The Lustgarten Foundation. The Pancreatic Cancer Collective 2021. Available from: https://pancreaticcancercollective.org/. |
| 25. | National Institutes of Health. Early Detection Research Network 2021. Available from: https://edrn.nci.nih.gov/. |
| 26. | Nakaura T, Higaki T, Awai K, Ikeda O, Yamashita Y. A primer for understanding radiology articles about machine learning and deep learning. Diagn Interv Imaging. 2020;101:765-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 27. | Nakata N. Recent technical development of artificial intelligence for diagnostic medical imaging. Jpn J Radiol. 2019;37:103-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 28. | Shalev-Shwartz S, Ben-David S. Understanding machine learning. From theory to algorithms. Understanding Machine Learning: From Theory to Algorithms. 2013. [DOI] [Full Text] |
| 29. | van der Sommen F, de Groof J, Struyvenberg M, van der Putten J, Boers T, Fockens K, Schoon EJ, Curvers W, de With P, Mori Y, Byrne M, Bergman JJGHM. Machine learning in GI endoscopy: practical guidance in how to interpret a novel field. Gut. 2020;69:2035-2045. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 30. | Kenner BJ, Abrams ND, Chari ST, Field BF, Goldberg AE, Hoos WA, Klimstra DS, Rothschild LJ, Srivastava S, Young MR, Go VLW. Early Detection of Pancreatic Cancer: Applying Artificial Intelligence to Electronic Health Records. Pancreas. 2021;50:916-922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 31. | Poruk KE, Firpo MA, Mulvihill SJ. Screening for pancreatic cancer. Adv Surg. 2014;48:115-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 32. | US Preventive Services Task Force, Owens DK, Davidson KW, Krist AH, Barry MJ, Cabana M, Caughey AB, Curry SJ, Doubeni CA, Epling JW Jr, Kubik M, Landefeld CS, Mangione CM, Pbert L, Silverstein M, Simon MA, Tseng CW, Wong JB. Screening for Pancreatic Cancer: US Preventive Services Task Force Reaffirmation Recommendation Statement. JAMA. 2019;322:438-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 239] [Article Influence: 39.8] [Reference Citation Analysis (1)] |
| 33. | Malhotra A, Rachet B, Bonaventure A, Pereira SP, Woods LM. Can we screen for pancreatic cancer? PLoS One. 2021;16:e0251876. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 34. | Chen Q, Cherry DR, Nalawade V, Qiao EM, Kumar A, Lowy AM, Simpson DR, Murphy JD. Clinical Data Prediction Model to Identify Patients With Early-Stage Pancreatic Cancer. JCO Clin Cancer Inform. 2021;5:279-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 35. | Appelbaum L, Cambronero JP, Stevens JP, Horng S, Pollick K, Silva G, Haneuse S, Piatkowski G, Benhaga N, Duey S, Stevenson MA, Mamon H, Kaplan ID, Rinard MC. Development and validation of a pancreatic cancer risk model for the general population using electronic health records: An observational study. Eur J Cancer. 2021;143:19-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 36. | Appelbaum L, Berg A, Cambronero JP, Dang THY, Jin CC, Zhang L, Kundrot S, Palchuk M, EvansLA, KaplanID, Rinard M. Development of a pancreatic cancer prediction model using a multinational medical records database. JCO. 2021;39:394. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 37. | Muhammad W, Hart GR, Nartowt B, Farrell JJ, Johung K, Liang Y, Deng J. Pancreatic Cancer Prediction Through an Artificial Neural Network. Front Artif Intell. 2019;2:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 38. | Placido D, Yuan B, Hjaltelin JX, Haue AD, Yuan C, Kim J, Umeton R, Antell G, Chowdhury A, Franz A, Brais L, Andrews E, Regev A, Kraft P, WolpinBM, Rosenthal M, Brunak S, Sander C. Pancreatic cancer risk predicted from disease trajectories using deep learning. BioRxiv. 2021;. [DOI] [Full Text] |
| 39. | Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, Kohane I. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc. 2010;17:124-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 554] [Cited by in RCA: 653] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 40. | Informatics OHDSA. Data Standardization 2021. Available from: https://ohdsi.org/data-standardization/. |
| 41. | Network TNP-CCR. Accelerating Data Value Across a National Community Health Center (ADVANCE) Network 2021. Available from: http://advancecollaborative.org/. |
| 42. | Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC Jr; ASCO. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313-5327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1110] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 43. | Sekiguchi M, Matsuda T. Limited usefulness of serum carcinoembryonic antigen and carbohydrate antigen 19-9 levels for gastrointestinal and whole-body cancer screening. Sci Rep. 2020;10:18202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 44. | Kriz D, Ansari D, Andersson R. Potential biomarkers for early detection of pancreatic ductal adenocarcinoma. Clin Transl Oncol. 2020;22:2170-2174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 45. | Brezgyte G, Shah V, Jach D, Crnogorac-Jurcevic T. Non-Invasive Biomarkers for Earlier Detection of Pancreatic Cancer-A Comprehensive Review. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 46. | Liao Q, Zhao YP, Yang YC, Li LJ, Long X, Han SM. Combined detection of serum tumor markers for differential diagnosis of solid lesions located at the pancreatic head. Hepatobiliary Pancreat Dis Int. 2007;6:641-645. [PubMed] |
| 47. | Yang Y, Chen H, Wang D, Luo W, Zhu B, Zhang Z. Diagnosis of pancreatic carcinoma based on combined measurement of multiple serum tumor markers using artificial neural network analysis. Chin Med J (Engl). 2014;127:1891-1896. [PubMed] |
| 48. | Yu J, Ploner A, Kordes M, Löhr M, Nilsson M, de Maturana MEL, Estudillo L, Renz H, Carrato A, Molero X, Real FX, Malats N, Ye W. Plasma protein biomarkers for early detection of pancreatic ductal adenocarcinoma. Int J Cancer. 2021;148:2048-2058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 49. | Ray K. Biomarkers for the early detection of PDAC. Nat Rev Gastroenterol Hepatol. 2017;14:505. |
| 50. | Young MR, Wagner PD, Ghosh S, Rinaudo JA, Baker SG, Zaret KS, Goggins M, Srivastava S. Validation of Biomarkers for Early Detection of Pancreatic Cancer: Summary of The Alliance of Pancreatic Cancer Consortia for Biomarkers for Early Detection Workshop. Pancreas. 2018;47:135-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 51. | Hasan S, Jacob R, Manne U, Paluri R. Advances in pancreatic cancer biomarkers. Oncol Rev. 2019;13:410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 52. | Tarasiuk A, Mackiewicz T, Małecka-Panas E, Fichna J. Biomarkers for early detection of pancreatic cancer - miRNAs as a potential diagnostic and therapeutic tool? Cancer Biol Ther. 2021;22:347-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 53. | Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, Calatayud D, Nielsen SE, Yilmaz M, Holländer NH, Andersen KK, Johansen JS. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311:392-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 342] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 54. | Duell EJ, Lujan-Barroso L, Sala N, Deitz McElyea S, Overvad K, Tjonneland A, Olsen A, Weiderpass E, Busund LT, Moi L, Muller D, Vineis P, Aune D, Matullo G, Naccarati A, Panico S, Tagliabue G, Tumino R, Palli D, Kaaks R, Katzke VA, Boeing H, Bueno-de-Mesquita HBA, Peeters PH, Trichopoulou A, Lagiou P, Kotanidou A, Travis RC, Wareham N, Khaw KT, Ramon Quiros J, Rodríguez-Barranco M, Dorronsoro M, Chirlaque MD, Ardanaz E, Severi G, Boutron-Ruault MC, Rebours V, Brennan P, Gunter M, Scelo G, Cote G, Sherman S, Korc M. Plasma microRNAs as biomarkers of pancreatic cancer risk in a prospective cohort study. Int J Cancer. 2017;141:905-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 55. | Khan IA, Rashid S, Singh N, Singh V, Gunjan D, Das P, Dash NR, Pandey RM, Chauhan SS, Gupta S, Saraya A. Panel of serum miRNAs as potential non-invasive biomarkers for pancreatic ductal adenocarcinoma. Sci Rep. 2021;11:2824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 56. | Shams R, Saberi S, Zali M, Sadeghi A, Ghafouri-Fard S, Aghdaei HA. Identification of potential microRNA panels for pancreatic cancer diagnosis using microarray datasets and bioinformatics methods. Sci Rep. 2020;10:7559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 57. | Cao Z, Liu C, Xu J, You L, Wang C, Lou W, Sun B, Miao Y, Liu X, Wang X, Zhang T, Zhao Y. Plasma microRNA panels to diagnose pancreatic cancer: Results from a multicenter study. Oncotarget. 2016;7:41575-41583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 58. | Almeida PP, Cardoso CP, de Freitas LM. PDAC-ANN: an artificial neural network to predict pancreatic ductal adenocarcinoma based on gene expression. BMC Cancer. 2020;20:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 59. | Alizadeh Savareh B, Asadzadeh Aghdaie H, Behmanesh A, Bashiri A, Sadeghi A, Zali M, Shams R. A machine learning approach identified a diagnostic model for pancreatic cancer through using circulating microRNA signatures. Pancreatology. 2020;20:1195-1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 60. | Yang J, Xu R, Wang C, Qiu J, Ren B, You L. Early screening and diagnosis strategies of pancreatic cancer: a comprehensive review. Cancer Commun (Lond). 2021;41:1257-1274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 166] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 61. | Khatri I, Bhasin MK. A Transcriptomics-Based Meta-Analysis Combined With Machine Learning Identifies a Secretory Biomarker Panel for Diagnosis of Pancreatic Adenocarcinoma. Front Genet. 2020;11:572284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 62. | Chung WY, Correa E, Yoshimura K, Chang MC, Dennison A, Takeda S, Chang YT. Using probe electrospray ionization mass spectrometry and machine learning for detecting pancreatic cancer with high performance. Am J Transl Res. 2020;12:171-179. [PubMed] |
| 63. | Ma H, Liu ZX, Zhang JJ, Wu FT, Xu CF, Shen Z, Yu CH, Li YM. Construction of a convolutional neural network classifier developed by computed tomography images for pancreatic cancer diagnosis. World J Gastroenterol. 2020;26:5156-5168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 56] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (1)] |
| 64. | Park S, Chu LC, Hruban RH, Vogelstein B, Kinzler KW, Yuille AL, Fouladi DF, Shayesteh S, Ghandili S, Wolfgang CL, Burkhart R, He J, Fishman EK, Kawamoto S. Differentiating autoimmune pancreatitis from pancreatic ductal adenocarcinoma with CT radiomics features. Diagn Interv Imaging. 2020;101:555-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 65. | Si K, Xue Y, Yu X, Zhu X, Li Q, Gong W, Liang T, Duan S. Fully end-to-end deep-learning-based diagnosis of pancreatic tumors. Theranostics. 2021;11:1982-1990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 66. | Watson MD, Lyman WB, Passeri MJ, Murphy KJ, Sarantou JP, Iannitti DA, Martinie JB, Vrochides D, Baker EH. Use of Artificial Intelligence Deep Learning to Determine the Malignant Potential of Pancreatic Cystic Neoplasms With Preoperative Computed Tomography Imaging. Am Surg. 2021;87:602-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 67. | Zhang Z, Li S, Wang Z, Lu Y. A Novel and Efficient Tumor Detection Framework for Pancreatic Cancer via CT Images. Annu Int Conf IEEE Eng Med Biol Soc. 2020;2020:1160-1164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 68. | Batts KP, Ludwig J. Chronic hepatitis. An update on terminology and reporting. Am J Surg Pathol. 1995;19:1409-1417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 823] [Cited by in RCA: 847] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 69. | Liu KL, Wu T, Chen PT, Tsai YM, Roth H, Wu MS, Liao WC, Wang W. Deep learning to distinguish pancreatic cancer tissue from non-cancerous pancreatic tissue: a retrospective study with cross-racial external validation. Lancet Digit Health. 2020;2:e303-e313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 70. | Chu LC, Park S, Kawamoto S, Fouladi DF, Shayesteh S, Zinreich ES, Graves JS, Horton KM, Hruban RH, Yuille AL, Kinzler KW, Vogelstein B, Fishman EK. Utility of CT Radiomics Features in Differentiation of Pancreatic Ductal Adenocarcinoma From Normal Pancreatic Tissue. AJR Am J Roentgenol. 2019;213:349-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 127] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 71. | Gao X, Wang X. Performance of deep learning for differentiating pancreatic diseases on contrast-enhanced magnetic resonance imaging: A preliminary study. Diagn Interv Imaging. 2020;101:91-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 72. | Zhu Z, Xia Y, Xie L, Fishman EK, Yuille AL. Multi-Scale Coarse-to-Fine Segmentation for Screening Pancreatic Ductal Adenocarcinoma. MICCAI; 2019. |
| 73. | Liu SL, Li S, Guo YT, Zhou YP, Zhang ZD, Lu Y. Establishment and application of an artificial intelligence diagnosis system for pancreatic cancer with a faster region-based convolutional neural network. Chin Med J (Engl). 2019;132:2795-2803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 74. | Chu LC, Park S, Kawamoto S, Wang Y, Zhou Y, Shen W, Zhu Z, Xia Y, Xie L, Liu F, Yu Q, Fouladi DF, Shayesteh S, Zinreich E, Graves JS, Horton KM, Yuille AL, Hruban RH, Kinzler KW, Vogelstein B, Fishman EK. Application of Deep Learning to Pancreatic Cancer Detection: Lessons Learned From Our Initial Experience. J Am Coll Radiol. 2019;16:1338-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 75. | Fu M, Wu W, Hong X, Liu Q, Jiang J, Ou Y, Zhao Y, Gong X. Hierarchical combinatorial deep learning architecture for pancreas segmentation of medical computed tomography cancer images. BMC Syst Biol. 2018;12:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 76. | Angulo P, Hui JM, Marchesini G, Bugianesi E, George J, Farrell GC, Enders F, Saksena S, Burt AD, Bida JP, Lindor K, Sanderson SO, Lenzi M, Adams LA, Kench J, Therneau TM, Day CP. The NAFLD fibrosis score: a noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology. 2007;45:846-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1917] [Cited by in RCA: 2276] [Article Influence: 126.4] [Reference Citation Analysis (1)] |
| 77. | Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, Obata M, Kurita Y, Koda H, Toriyama K, Onishi S, Ishihara M, Tanaka T, Tajika M, Niwa Y. Usefulness of Deep Learning Analysis for the Diagnosis of Malignancy in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin Transl Gastroenterol. 2019;10:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 126] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 78. | Marya NB, Powers PD, Chari ST, Gleeson FC, Leggett CL, Abu Dayyeh BK, Chandrasekhara V, Iyer PG, Majumder S, Pearson RK, Petersen BT, Rajan E, Sawas T, Storm AC, Vege SS, Chen S, Long Z, Hough DM, Mara K, Levy MJ. Utilisation of artificial intelligence for the development of an EUS-convolutional neural network model trained to enhance the diagnosis of autoimmune pancreatitis. Gut. 2021;70:1335-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 85] [Article Influence: 21.3] [Reference Citation Analysis (1)] |
| 79. | Norton ID, Zheng Y, Wiersema MS, Greenleaf J, Clain JE, Dimagno EP. Neural network analysis of EUS images to differentiate between pancreatic malignancy and pancreatitis. Gastrointest Endosc. 2001;54:625-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 80. | Ozkan M, Cakiroglu M, Kocaman O, Kurt M, Yilmaz B, Can G, Korkmaz U, Dandil E, Eksi Z. Age-based computer-aided diagnosis approach for pancreatic cancer on endoscopic ultrasound images. Endosc Ultrasound. 2016;5:101-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 81. | Săftoiu A, Vilmann P, Dietrich CF, Iglesias-Garcia J, Hocke M, Seicean A, Ignee A, Hassan H, Streba CT, Ioncică AM, Gheonea DI, Ciurea T. Quantitative contrast-enhanced harmonic EUS in differential diagnosis of focal pancreatic masses (with videos). Gastrointest Endosc. 2015;82:59-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 112] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 82. | Săftoiu A, Vilmann P, Gorunescu F, Janssen J, Hocke M, Larsen M, Iglesias-Garcia J, Arcidiacono P, Will U, Giovannini M, Dietrich CF, Havre R, Gheorghe C, McKay C, Gheonea DI, Ciurea T; European EUS Elastography Multicentric Study Group. Efficacy of an artificial neural network-based approach to endoscopic ultrasound elastography in diagnosis of focal pancreatic masses. Clin Gastroenterol Hepatol. 2012;10:84-90.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 116] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 83. | Tonozuka R, Itoi T, Nagata N, Kojima H, Sofuni A, Tsuchiya T, Ishii K, Tanaka R, Nagakawa Y, Mukai S. Deep learning analysis for the detection of pancreatic cancer on endosonographic images: a pilot study. J Hepatobiliary Pancreat Sci. 2021;28:95-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 84. | Udriștoiu AL, Cazacu IM, Gruionu LG, Gruionu G, Iacob AV, Burtea DE, Ungureanu BS, Costache MI, Constantin A, Popescu CF, Udriștoiu Ș, Săftoiu A. Real-time computer-aided diagnosis of focal pancreatic masses from endoscopic ultrasound imaging based on a hybrid convolutional and long short-term memory neural network model. PLoS One. 2021;16:e0251701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 85. | Zhang MM, Yang H, Jin ZD, Yu JG, Cai ZY, Li ZS. Differential diagnosis of pancreatic cancer from normal tissue with digital imaging processing and pattern recognition based on a support vector machine of EUS images. Gastrointest Endosc. 2010;72:978-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 86. | Zhu M, Xu C, Yu J, Wu Y, Li C, Zhang M, Jin Z, Li Z. Differentiation of pancreatic cancer and chronic pancreatitis using computer-aided diagnosis of endoscopic ultrasound (EUS) images: a diagnostic test. PLoS One. 2013;8:e63820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (2)] |
| 87. | Singh DP, Sheedy S, Goenka AH, Wells M, Lee NJ, Barlow J, Sharma A, Kandlakunta H, Chandra S, Garg SK, Majumder S, Levy MJ, Takahashi N, Chari ST. Computerized tomography scan in pre-diagnostic pancreatic ductal adenocarcinoma: Stages of progression and potential benefits of early intervention: A retrospective study. Pancreatology. 2020;20:1495-1501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 88. | Yu J, Blackford AL, Dal Molin M, Wolfgang CL, Goggins M. Time to progression of pancreatic ductal adenocarcinoma from low-to-high tumour stages. Gut. 2015;64:1783-1789. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 169] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 89. | Overbeek KA, Goggins MG, Dbouk M, Levink IJM, Koopmann BDM, Chuidian M, Konings ICAW, Paiella S, Earl J, Fockens P, Gress TM, Ausems MGEM, Poley JW, Thosani NC, Half E, Lachter J, Stoffel EM, Kwon RS, Stoita A, Kastrinos F, Lucas AL, Syngal S, Brand RE, Chak A, Carrato A, Vleggaar FP, Bartsch DK, van Hooft JE, Cahen DL, Canto MI, Bruno MJ; International Cancer of the Pancreas Screening Consortium. Timeline of Development of Pancreatic Cancer and Implications for Successful Early Detection in High-Risk Individuals. Gastroenterology. 2022;162:772-785.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 79] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 90. | Young MR, Abrams N, Ghosh S, Rinaudo JAS, Marquez G, Srivastava S. Prediagnostic Image Data, Artificial Intelligence, and Pancreatic Cancer: A Tell-Tale Sign to Early Detection. Pancreas. 2020;49:882-886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 91. | Springer S, Masica DL, Dal Molin M, Douville C, Thoburn CJ, Afsari B, Li L, Cohen JD, Thompson E, Allen PJ, Klimstra DS, Schattner MA, Schmidt CM, Yip-Schneider M, Simpson RE, Fernandez-Del Castillo C, Mino-Kenudson M, Brugge W, Brand RE, Singhi AD, Scarpa A, Lawlor R, Salvia R, Zamboni G, Hong SM, Hwang DW, Jang JY, Kwon W, Swan N, Geoghegan J, Falconi M, Crippa S, Doglioni C, Paulino J, Schulick RD, Edil BH, Park W, Yachida S, Hijioka S, van Hooft J, He J, Weiss MJ, Burkhart R, Makary M, Canto MI, Goggins MG, Ptak J, Dobbyn L, Schaefer J, Sillman N, Popoli M, Klein AP, Tomasetti C, Karchin R, Papadopoulos N, Kinzler KW, Vogelstein B, Wolfgang CL, Hruban RH, Lennon AM. A multimodality test to guide the management of patients with a pancreatic cyst. Sci Transl Med. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 92. | Yang Z, Zhang L, Zhang M, Feng J, Wu Z, Ren F, Lv Y. Pancreas Segmentation in Abdominal CT Scans using Inter-/Intra-Slice Contextual Information with a Cascade Neural Network. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:5937-5940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 93. | Barat M, Chassagnon G, Dohan A, Gaujoux S, Coriat R, Hoeffel C, Cassinotto C, Soyer P. Artificial intelligence: a critical review of current applications in pancreatic imaging. Jpn J Radiol. 2021;39:514-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |