Published online Aug 8, 2024. doi: 10.35712/aig.v5.i2.91550
Revised: July 6, 2024
Accepted: July 29, 2024
Published online: August 8, 2024
Processing time: 190 Days and 21.6 Hours
Digital pathology (DP) and its subsidiaries including artificial intelligence (AI) are rapidly making inroads into the area of diagnostic anatomic pathology (AP) including gastrointestinal (GI) pathology. It is poised to revolutionize the field of diagnostic AP. Historically, AP has been slow to adopt digital technology, but this is changing rapidly, with many centers worldwide transitioning to DP. Coupled with advanced techniques of AI such as deep learning and machine learning, DP is likely to transform histopathology from a subjective field to an objective, efficient, and transparent discipline. AI is increasingly integrated into GI patho
Core Tip: Anatomic pathology remains largely subjective compared to other diagnostic laboratory fields. However, the digitization of tissue sections and the development of artificial intelligence-based technologies are rapidly advancing image-based diagnostics in anatomic pathology including gastrointestinal pathology. These technologies allow pathologists to make diagnoses more quickly and accurately, particularly for time-consuming and repetitive tasks, leading to higher volumes and faster turnaround times. Increasing awareness of the potential uses and benefits of these emerging technologies is essential for the pathology community.
- Citation: Mubarak M, Rashid R, Sapna F, Shakeel S. Expanding role and scope of artificial intelligence in the field of gastrointestinal pathology. Artif Intell Gastroenterol 2024; 5(2): 91550
- URL: https://www.wjgnet.com/2644-3236/full/v5/i2/91550.htm
- DOI: https://dx.doi.org/10.35712/aig.v5.i2.91550
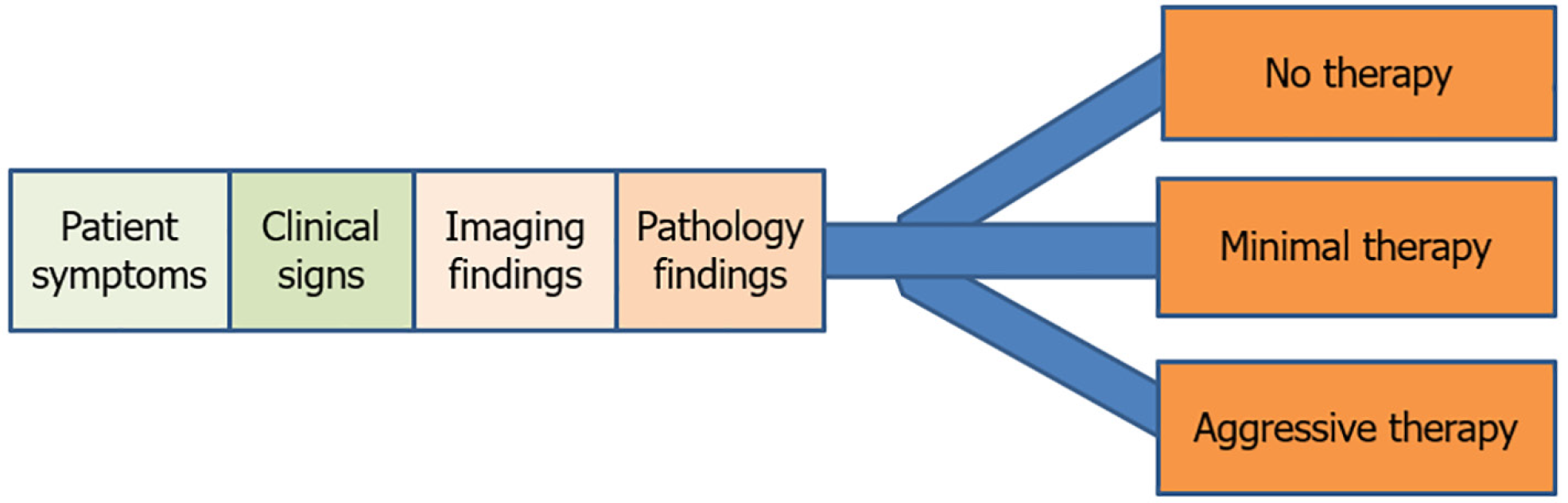
Anatomic pathology (AP), particularly histopathology, represents the ground truth of medicine, providing the final and definitive test on which crucial treatment decisions are based, especially for cancer (Figure 1). Despite its critical role, AP has remained an analog enterprise, using processes developed in the early 20th century. Tissue preparation and diagnosis are still largely manual and subjective[1,2]. Diagnoses are based on the visualization and assessment of tissue sections on glass slides under a light microscope, making the process highly dependent on the pathologist’s interpretation.
The diagnostic process is a complex mental exercise requiring multitasking and the coordination of observation, interpretation, and integration of information. This process yields continuous variables that pathologists use to drive classification systems, which clinicians use to make major therapeutic decisions (Figure 1). While cost-effective, this process is prone to significant inter- and intra-pathologist variation and diagnostic errors and is often time-consuming and tedious. The integration of multiple ancillary diagnostic tests, such as immunohistochemistry and molecular assays, adds to the complexity and demands on pathologists. Moreover, the shortage of pathologists globally exacerbates these challenges, highlighting the need for precision diagnostics, particularly in cancer treatment[3-5].
Traditionally, AP has been slow to embrace digital technology, but this is steadily changing. Many pathology laboratories worldwide have partially or completely transitioned to digital pathology (DP) workflows[6-10]. Advanced artificial intelligence (AI) techniques, such as machine learning (ML) and deep learning (DL), are set to transform AP into a more objective, efficient, and transparent discipline[11,12]. However, many pathologists, particularly in developing countries, have limited knowledge of AI and its vast potential[13,14].
This review aimed to provide a simplified overview of the latest developments in the role and scope of AI in pathology, with a specific focus on gastrointestinal (GI) pathology. It explored how AI can be used to diagnose and predict diseases, highlighting its benefits for routine histopathology practice. The goal was to update pathologists and other healthcare providers about these emerging diagnostic technologies and raise awareness.
A comprehensive search was conducted across multiple databases, including PubMed, Google Scholar, Scopus, and Web of Science, covering publications from 2010 to 2024. Keywords included “Artificial Intelligence,” “Machine Learning,” “Deep Learning,” “Gastrointestinal Pathology,” “Diagnosis,” and “Histopathology.” All types of studies in the English language were selected based on relevance, focusing on AI applications in GI pathology. The full texts of these articles were carefully read to extract the relevant points for writing this review article.
AI is a field of computer science that enables computers to perform tasks that typically require human intelligence, such as learning, pattern recognition, planning, problem-solving, and reasoning[15-17]. AI relies heavily on data for its operations, and the digitization of glass slides in DP workflows provides vast amounts of pixel data for AI applications. AI can be considered a part of data science[18-21].
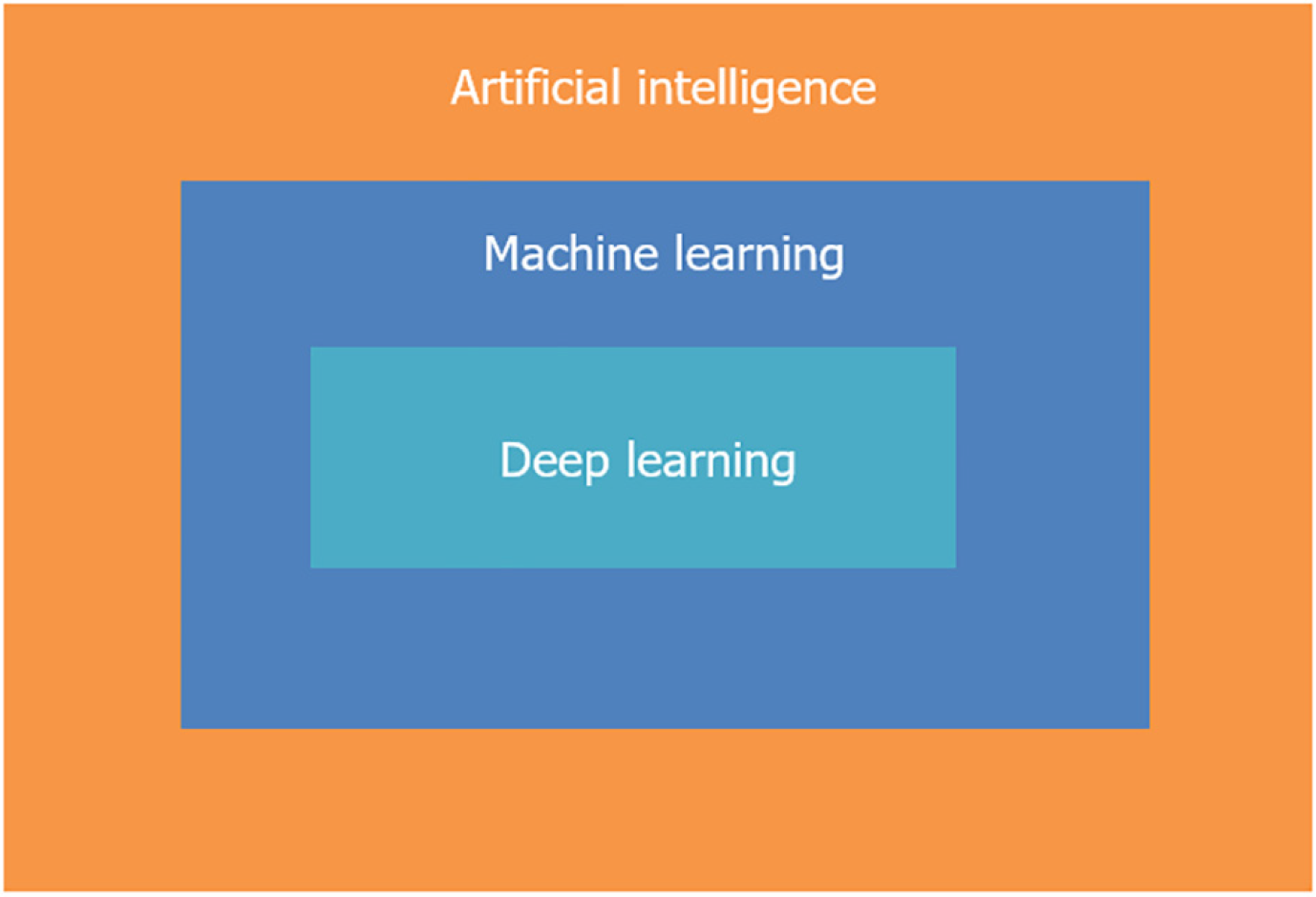
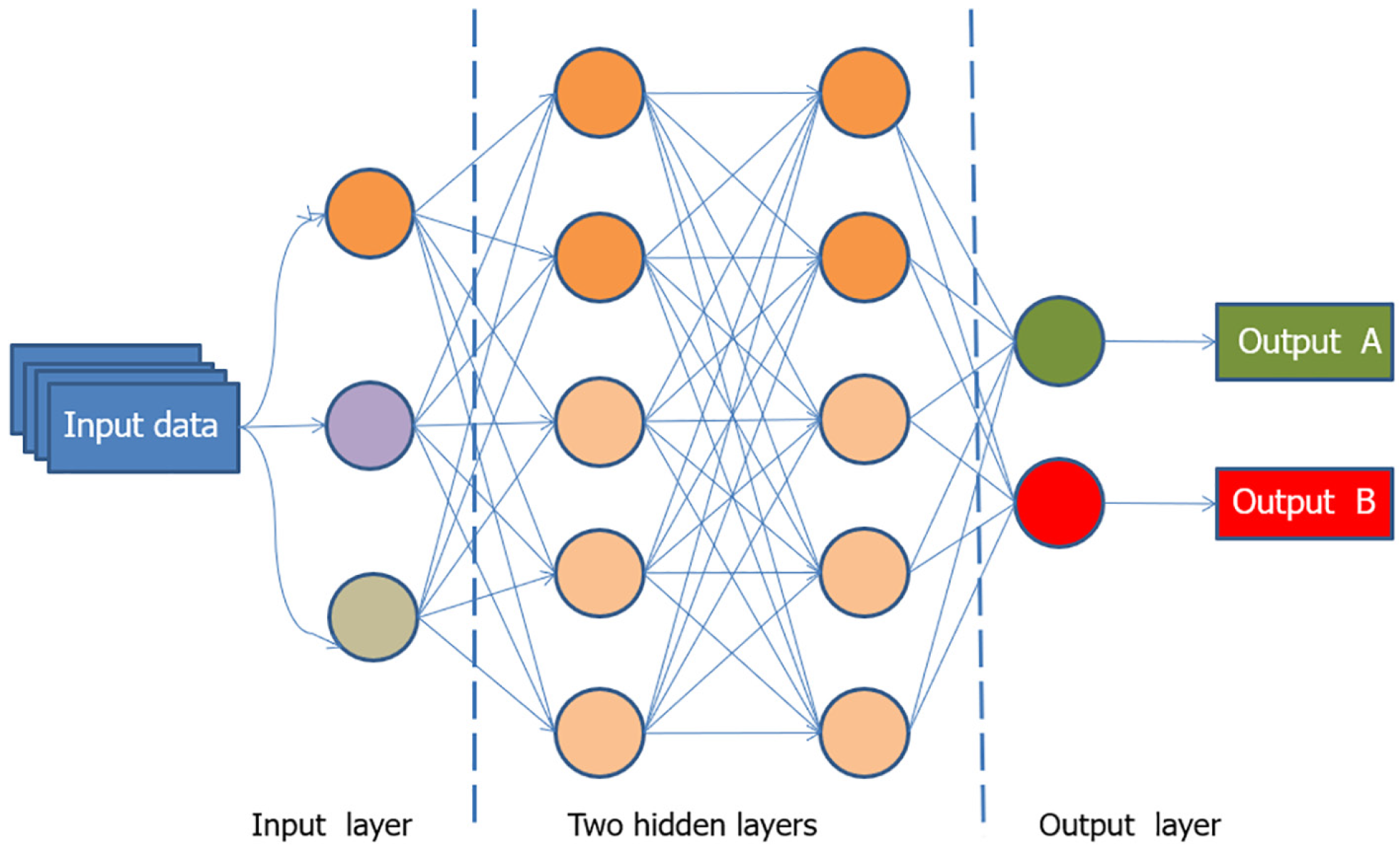
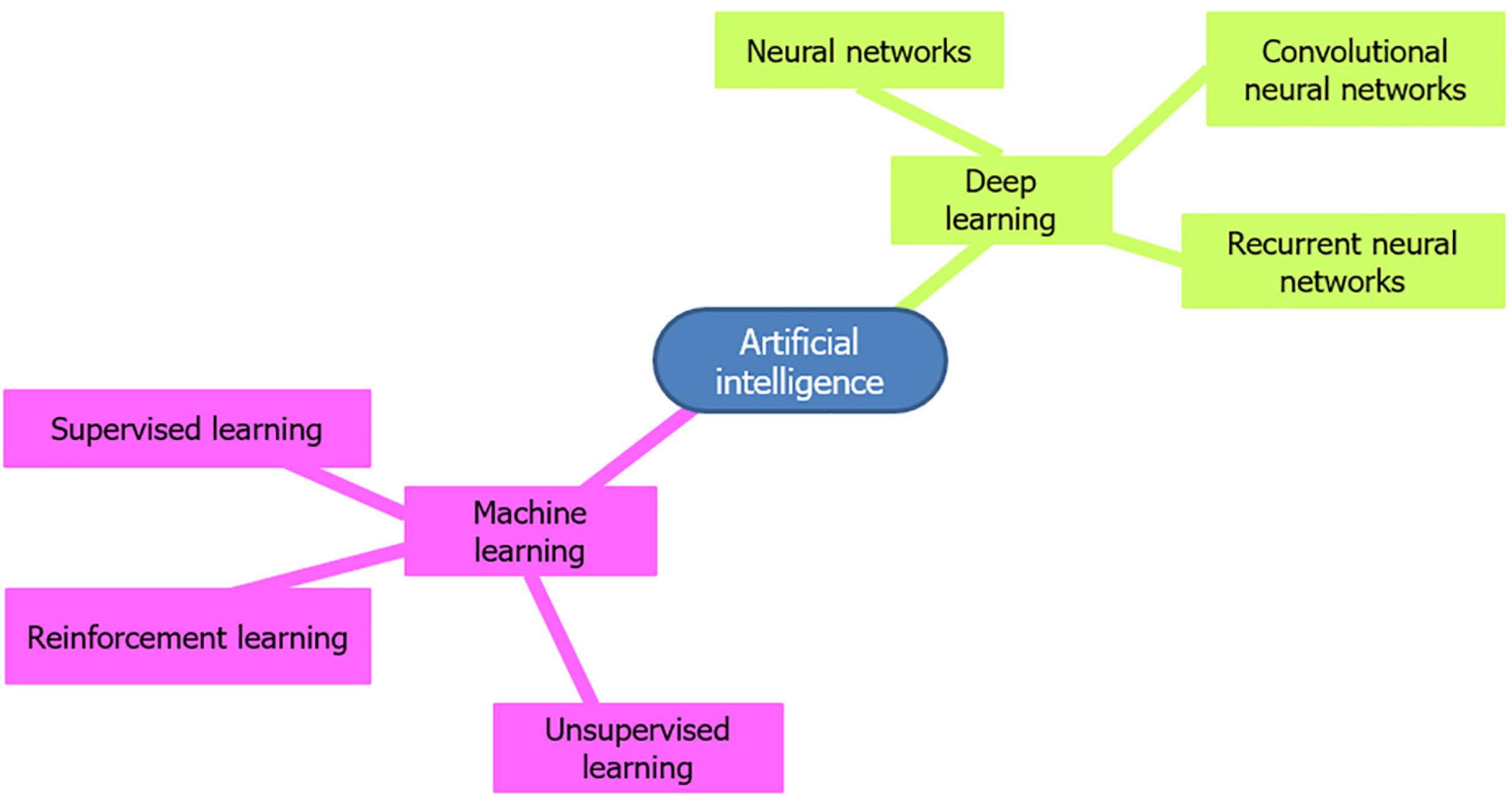
Initially, AI comprised simple “if-then” algorithms but has since advanced to include complex algorithms that perform tasks akin to the human brain. The advent of DL has expanded the capabilities of AI, allowing systems to analyze data and images using multiple layers and learn from big data. ML and DL are subsets of AI (Figure 2), with DL being a more advanced form capable of solving complex problems using neural networks (Figure 3)[22-24].
AI represents a significant turning point for human society, comparable to the industrial revolution. It is a general purpose technology applicable in various fields, much like electricity. The role of AI in healthcare is growing rapidly, particularly in biomedical research and clinical practice. Modern techniques now generate vast amounts of data, which AI can analyze for patterns, enhancing diagnostics and treatment decisions[25-29]. AI can detect subtle pathological alterations, predict therapy responses, and improve workflow efficiency in pathology[30].
In the diagnostics arena, radiologists have been early adopters of AI for image processing and interpretation[31,32]. Pathologists, facing greater visual data complexity in microscopic images, have been slower to adopt AI. However, the shift towards DP and whole slide imaging (WSI) is laying the groundwork for computational pathology technologies. With the recent advancements in AI for computer vision, it is expected that AI will soon support pathologists in various DP tasks. Concurrently, significant progress in DL has created a synergy with AI, enabling image-based diagnostics within the DP context. Efforts are underway to develop AI tools that save pathologists time and reduce errors[33]. Integrating AI systems into AP practices will require fully digital imaging platforms, updating outdated information technology infrastructures, modifying laboratory and pathologist workflows, establishing appropriate reimbursement models, and ensuring pathologists’ active participation for buy-in and oversight. New regulations, designed to address the unique aspects and limitations of AI, are being developed to ensure its safe and effective use[34]. The recent Food and Drug Administration approval of WSI systems opens significant opportunities for AI-assisted pathological diagnosis, promising faster, more accurate, and cost-effective diagnostics[35-40].
AI is increasingly being integrated into AP to enhance diagnostic efficiency and accuracy, reduce turnaround times, and improve patient care[41,42]. AI algorithms can analyze DP images with high speed and accuracy, assisting pathologists in identifying and quantifying specific features such as cell structures, mitoses, tissue patterns, and abnormalities. This reduces subjectivity, minimizes diagnostic errors, and ensures consistent results[43-47].
AI-driven workflow management tools can streamline daily tasks, prioritize cases based on urgency, and help pathologists allocate their time effectively. AI can also integrate patient data, provide decision support tools, and assist in quality control and compliance with regulatory standards. AI tools can develop predictive models for disease outcomes and support research by analyzing vast datasets[48,49]. Although technical implementation has become less challenging, much work is needed to integrate AI into routine AP workflows. AI can also enhance the understanding of disease biology by analyzing DP images to identify patterns and features not visible to the human eye. This can aid in disco
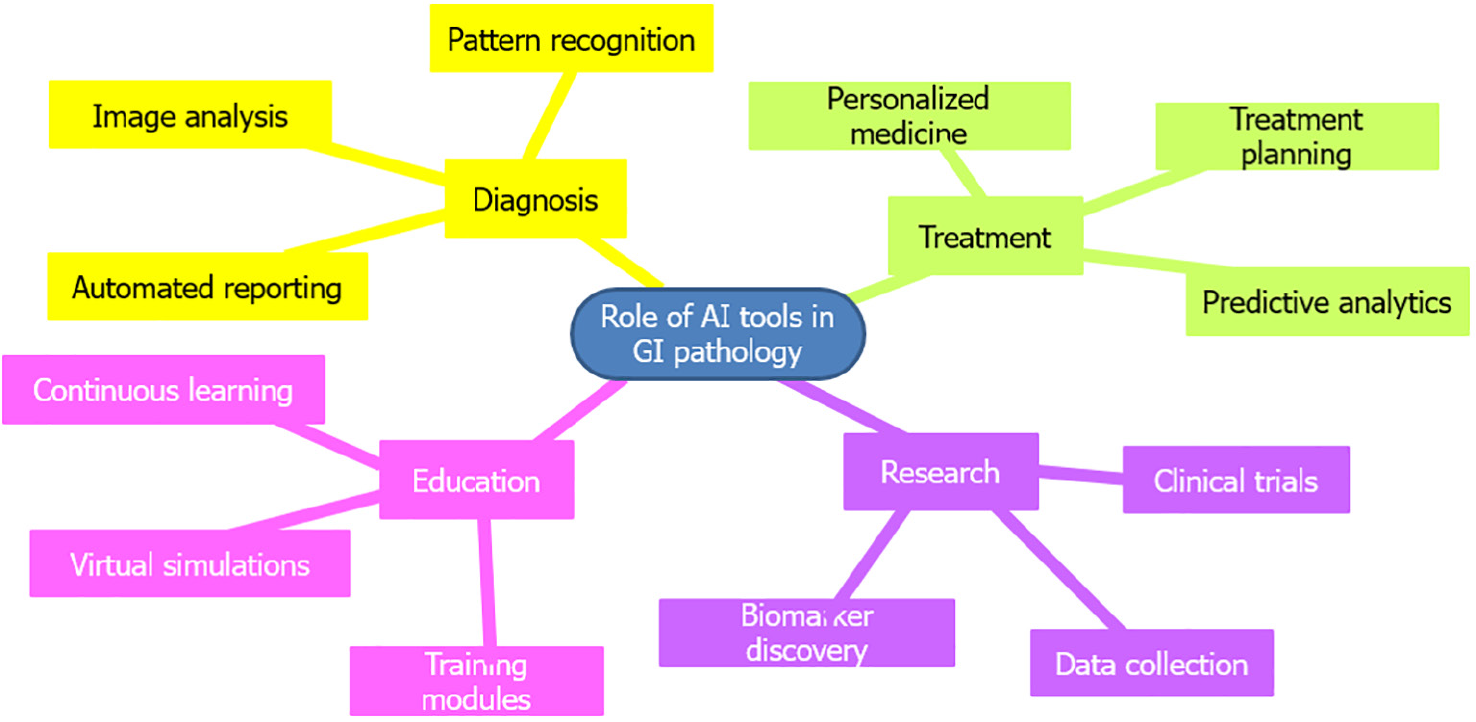
AI technology is poised to revolutionize GI pathology, offering numerous current and potential applications (Figure 4). By processing digitized images of tissue samples, AI tools enhance the precise and effective identification of various GI disease processes, including inflammatory and neoplastic conditions such as colitis, Crohn’s disease, and colorectal cancer (CRC). The integration of AI in GI pathology significantly improves the precision, speed, and quality of diagnostic and therapeutic decision-making processes, ultimately benefiting patient care. Additionally, AI can standardize quality control in GI pathology, ensuring accurate and consistent results across samples[52-55].
AI has been extensively studied for endoscopic diagnosis of GI tract disorders, demonstrating significant promise. It is expected that AI will primarily assist endoscopists with tasks such as detection, characterization, and segmentation. AI has the potential to enhance colonoscopy-based colorectal screening and monitoring by reducing unnecessary expenses and improving quality. Real-time computer-assisted polyp identification can enhance screening and monitoring quality, as measured by adenoma detection rates. Optical biopsies using computer-assisted diagnosis can identify low-risk polyps, supporting resect-and-discard or diagnose-and-leave strategies, thereby reducing unnecessary costs. Recent meta-analyses indicated that AI tools significantly increased colorectal neoplasia detection, regardless of initial adenoma features[56-58]. Furthermore, AI is useful in identifying upper GI pathological processes, including both neoplastic and non-neoplastic lesions[59,60].
In the GI tract, precancerous lesions and invasive tumors are routinely biopsied or excised for histopathological workup. Early and accurate diagnosis is a primary responsibility of pathologists, and AI can assist in achieving this objective. Numerous reports have documented AI-assisted diagnosis of both neoplastic and non-neoplastic GI diseases. For instance, Korbar et al[61] trained a model to distinguish between five prevalent types of colorectal polyps with an overall accuracy of 93% using a dataset of over 400 WSIs. Wei et al[62] demonstrated that neural networks trained to identify colorectal polyps on WSIs from one institution performed similarly to local pathologists when applied to WSIs from other institutions. Efforts have also been made to automate the diagnosis of preneoplastic and neoplastic lesions, such as Barrett’s esophagus or gastric adenomas/adenocarcinomas[60].
AI models also show promise in predicting therapy response or prognosis from WSI analysis. Among all cancer types, GI and liver tumors have notably driven computational oncology forward. AI can extract complex information from digital images of GI and hepatic malignancies, providing clinical, biological, and molecular insights that are not accessible to the naked eye. By identifying the most predictive tissue areas, AI reduces the cognitive burden on pathologists, enhancing their efficacy in histopathological characterization and risk assessments of GI preneoplastic and neoplastic lesions. In biliary tract cancer, DL can identify tissue features predictive of clinical outcomes. DP images and tissue microarrays from CRC have shown the efficacy of DL in prognostic prediction across all tumor stages. The histomorphology of gastric cancer (GC) is more complex and variable than CRC, leading to fewer investigations using DL for GC. Most of this research has focused on tumor detection rather than prognostication[63-66].
Routine processing of surgical and biopsy specimens from various GI tract tumors involves investigating molecular biomarkers that predict responses to targeted therapy. Specific genetic events in GI and hepatobiliary cancers are linked to morphological features identified in hematoxylin and eosin sections. AI-based algorithms on WSIs have been successfully used as surrogate markers for these alterations[66-69]. For example, CRC serves as a model disease due to the abundance of pathology samples. Identifying microsatellite instability (MSI) is crucial because immune-modulating treatments significantly affect MSI tumors. MSI identification has major implications for patients and their families, necessitating further investigation to identify Lynch syndrome. Although immunohistochemistry techniques are typically used to identify MSI, not all patients are routinely screened. A study by Echle et al[70] examined 8836 CRC cases across all stages, developing an AI model that could identify MSI tumors from hematoxylin and eosin sections, maintaining performance even in biopsy samples with limited tissue and varying preprocessing methods. Other efforts have created models that accurately predict gene alterations from WSIs of hepatocellular carcinoma (HCC), GC, and other conditions.
AI-based pathology can predict gene expression and RNA sequencing data, holding great promise for clinical application. Developing DL models for prognostication that integrate clinical, biological, and genetic data is a promising approach. For example, Chaudhary et al[71] used RNA sequencing, microRNA sequencing, and methylation data to create a DL model predicting survival in HCC patients, demonstrating its efficacy across different HCC patient cohorts.
AI algorithms, particularly those based on ML and DL, have shown substantial potential in analyzing complex pathological data and are central to the advancements in GI pathology (Table 1). ML algorithms such as support vector machines, random forests, and k-nearest neighbors have been employed to classify and predict various GI conditions. These include supervised, unsupervised, and reinforcement learning algorithms (Figure 5). Supervised learning algorithms, such as support vector machines and random forests, are widely used for classification tasks. These algorithms require extensive feature engineering and domain expertise to identify relevant features from pathology images and clinical data. They are relatively simpler to implement and interpret compared to DL algorithms. They are also effective for structured data analysis and smaller datasets and feature faster training times and lower computational requirements. The need for manual feature extraction limits their use, and ML algorithms may not perform as well as DL in recognizing complex patterns in unstructured data like histopathological images[72-75].
| AI algorithms | Role in gastrointestinal pathology | Key uses | Examples |
| Machine learning | Assisting in diagnosis and classification of gastrointestinal diseases | Improved diagnostic accuracy, personalized treatment plans | Predictive models for colorectal cancer risk, classification of polyps in colonoscopy images |
| Deep learning | Analyzing endoscopic and histopathologic images | Enhanced image recognition, reduced human error | Convolutional neural networks for detecting and classifying lesions in endoscopic images |
| Natural language processing | Extracting relevant information from medical records and literature | Efficient data mining, real-time clinical decision support | Automated extraction of patient data from electronic health records for research and clinical use |
| Computer vision | Real-time analysis of endoscopic videos | Immediate feedback during procedures, increased detection rates of abnormalities | Detection of bleeding, polyps, and other abnormalities during live endoscopy procedures |
| Reinforcement learning | Optimizing treatment plans and clinical pathways | Adaptive learning from real-world outcomes, improved clinical decision-making | Personalized treatment strategies for inflammatory bowel disease based on patient response |
| Predictive analytics | Forecasting disease progression and patient outcomes | Proactive patient management, early intervention | Predicting flare-ups in Crohn’s disease, forecasting outcomes after gastrointestinal surgeries |
| Robotics integration | Enhancing precision in minimally invasive surgeries | Increased surgical precision, reduced recovery time | AI-assisted robotic surgery for gastrointestinal procedures, such as robotic-assisted colectomy |
| Genomic data analysis | Identifying genetic markers associated with gastrointestinal diseases | Personalized medicine, targeted therapies | Analyzing genetic data to find markers for conditions like Lynch syndrome and hereditary pancreatitis |
DL models, particularly convolutional neural networks, have revolutionized image analysis in GI pathology[76,77]. Convolutional neural networks can automatically learn hierarchical features from raw images, making them highly effective for tasks such as tumor detection and classification. They possess superior performance in image recognition tasks and are able to handle large and complex datasets. Automated feature extraction reduces the need for domain-specific knowledge. However, it requires substantial computational resources and large annotated datasets. It is difficult to interpret and explain the decision-making process (black-box nature). Recurrent neural networks, including their variants like long short-term memory networks, are used for sequential data analysis. They are particularly useful in analyzing time-series data from endoscopic videos to detect abnormalities[78-84]. While ML algorithms are generally more interpretable, DL algorithms often provide higher accuracy due to their ability to learn complex patterns from large datasets. However, DL models require substantial computational resources and large labeled datasets, which can be a limitation[73,75,79,85].
The performance of AI algorithms heavily depends on the quality and diversity of data sources. Common data sources in GI pathology include: (1) Histopathological images in the form of WSIs and tissue microarrays as the primary data sources; (2) Clinical data, such as electronic health records, patient demographics, clinical history, and endoscopy images; and (3) Publicly available datasets such as The Cancer Genome Atlas and Gastrointestinal Image Data Collection. Each of these sources has merits and demerits. Integration of clinical data, including patient demographics, medical history, and laboratory results, enhances the contextual understanding of GI pathology and improves the predictive power of AI models. The availability of large, well-annotated datasets is a significant challenge. Variability in image quality and staining techniques and differences in pathological practices across institutions can affect the generalizability of AI models. Additionally, integrating clinical data requires sophisticated data management systems to handle patient privacy and data security concerns.
Sensitivity measures the ability of an AI algorithm to correctly identify positive cases (e.g., detecting cancerous lesions). Specificity measures the ability of an AI algorithm to correctly identify negative cases (e.g., ruling out benign conditions). Achieving high sensitivity and specificity is challenging due to the inherent variability in pathological samples. DL models often outperform traditional ML models in sensitivity and specificity due to their ability to learn intricate features from large datasets. However, there is a trade-off; models with high sensitivity might produce more false positives, reducing specificity. Balancing these metrics is essential to avoid misdiagnoses and unnecessary treatments[74,76,81,82].
AI is still in its early stages, and many pathology laboratories worldwide have yet to transition to a digital workflow to fully benefit from AI technologies. There are numerous obstacles to the widespread implementation of AI solutions in routine clinical practice, even in developed countries. Bringing an AI solution for pathology to market poses significant technological, business, and regulatory challenges. Although some clinical applications exist, the overall introduction of AI into medical practice has been slow and not without ethical concerns[86-90].
Despite significant research developments in AI-based techniques in recent years, only a few AI solutions have become commercial products for routine use. Consequently, much of the potential of AI remains untapped. Research models need further development, improvement, and integration into the information technology infrastructure of clinical laboratories before they can be used in routine pathology workflows. Additionally, commercial success requires a profitable business model in most countries, and pathologists need to be reimbursed for using the product. AI solutions are also classified as medical devices and thus require regulatory approval before they can be sold as products[91-94].
The role and scope of AI are expanding in GI pathology, with the potential to improve diagnostic accuracy, efficiency, and patient care. Increasing awareness among the pathology community about these emerging technologies is essential to realize their full potential and revolutionize diagnostics, prognostics, and theranostics in GI pathology.
| 1. | Mubarak M. From Digital to Computational Pathology and Integrated Diagnostics: The Future of Histopathology. J Coll Physicians Surg Pak. 2021;31:2-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 2. | Pantanowitz L, Sharma A, Carter AB, Kurc T, Sussman A, Saltz J. Twenty Years of Digital Pathology: An Overview of the Road Travelled, What is on the Horizon, and the Emergence of Vendor-Neutral Archives. J Pathol Inform. 2018;9:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 135] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 3. | Kumar N, Gupta R, Gupta S. Whole Slide Imaging (WSI) in Pathology: Current Perspectives and Future Directions. J Digit Imaging. 2020;33:1034-1040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 123] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 4. | Boyce BF. Whole slide imaging: uses and limitations for surgical pathology and teaching. Biotech Histochem. 2015;90:321-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 5. | Jahn SW, Plass M, Moinfar F. Digital Pathology: Advantages, Limitations and Emerging Perspectives. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 139] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 6. | Kiran N, Sapna F, Kiran F, Kumar D, Raja F, Shiwlani S, Paladini A, Sonam F, Bendari A, Perkash RS, Anjali F, Varrassi G. Digital Pathology: Transforming Diagnosis in the Digital Age. Cureus. 2023;15:e44620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 7. | Yilmaz F, Brickman A, Najdawi F, Yakirevich E, Egger R, Resnick MB. Advancing Artificial Intelligence Integration Into the Pathology Workflow: Exploring Opportunities in Gastrointestinal Tract Biopsies. Lab Invest. 2024;104:102043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 8. | Hanna MG, Parwani A, Sirintrapun SJ. Whole Slide Imaging: Technology and Applications. Adv Anat Pathol. 2020;27:251-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 9. | Iyengar JN. Whole slide imaging: The futurescape of histopathology. Indian J Pathol Microbiol. 2021;64:8-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 10. | Jain E, Patel A, Parwani AV, Shafi S, Brar Z, Sharma S, Mohanty SK. Whole Slide Imaging Technology and Its Applications: Current and Emerging Perspectives. Int J Surg Pathol. 2024;32:433-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 11. | Volynskaya Z, Evans AJ, Asa SL. Clinical Applications of Whole-slide Imaging in Anatomic Pathology. Adv Anat Pathol. 2017;24:215-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 12. | Pantanowitz L, Sinard JH, Henricks WH, Fatheree LA, Carter AB, Contis L, Beckwith BA, Evans AJ, Lal A, Parwani AV; College of American Pathologists Pathology and Laboratory Quality Center. Validating whole slide imaging for diagnostic purposes in pathology: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2013;137:1710-1722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 441] [Cited by in RCA: 429] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 13. | Zehra T, Parwani A, Abdul-Ghafar J, Ahmad Z. A suggested way forward for adoption of AI-Enabled digital pathology in low resource organizations in the developing world. Diagn Pathol. 2023;18:68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 14. | Mubarak M. Move from Traditional Histopathology to Digital and Computational Pathology: Are we Ready? Indian J Nephrol. 2022;32:414-415. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 15. | Schüffler P, Steiger K, Weichert W. How to use AI in pathology. Genes Chromosomes Cancer. 2023;62:564-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 858] [Cited by in RCA: 844] [Article Influence: 140.7] [Reference Citation Analysis (0)] |
| 17. | Ahmad Z, Rahim S, Zubair M, Abdul-Ghafar J. Artificial intelligence (AI) in medicine, current applications and future role with special emphasis on its potential and promise in pathology: present and future impact, obstacles including costs and acceptance among pathologists, practical and philosophical considerations. A comprehensive review. Diagn Pathol. 2021;16:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 97] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 18. | Yang YC, Islam SU, Noor A, Khan S, Afsar W, Nazir S. Influential Usage of Big Data and Artificial Intelligence in Healthcare. Comput Math Methods Med. 2021;2021:5812499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 19. | Försch S, Klauschen F, Hufnagl P, Roth W. Artificial Intelligence in Pathology. Dtsch Arztebl Int. 2021;118:194-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 20. | Berezowska S, Cathomas G, Grobholz R, Henkel M, Jochum W, Koelzer VH, Kreutzfeldt M, Mertz KD, Rössle M, Soldini D, Zlobec I, Janowczyk A. Digital image analysis and artificial intelligence in pathology diagnostics-the Swiss view. Pathologie (Heidelb). 2023;44:222-224. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 21. | Jiang Y, Yang M, Wang S, Li X, Sun Y. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun (Lond). 2020;40:154-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 241] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 22. | Serag A, Ion-Margineanu A, Qureshi H, McMillan R, Saint Martin MJ, Diamond J, O'Reilly P, Hamilton P. Translational AI and Deep Learning in Diagnostic Pathology. Front Med (Lausanne). 2019;6:185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 145] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 23. | Sultan AS, Elgharib MA, Tavares T, Jessri M, Basile JR. The use of artificial intelligence, machine learning and deep learning in oncologic histopathology. J Oral Pathol Med. 2020;49:849-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 85] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 24. | Sharma P, Hassan C. Artificial Intelligence and Deep Learning for Upper Gastrointestinal Neoplasia. Gastroenterology. 2022;162:1056-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 25. | Baxi V, Edwards R, Montalto M, Saha S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol. 2022;35:23-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 265] [Article Influence: 88.3] [Reference Citation Analysis (0)] |
| 26. | Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nat Med. 2022;28:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 930] [Article Influence: 310.0] [Reference Citation Analysis (0)] |
| 27. | Polevikov S. Advancing AI in healthcare: A comprehensive review of best practices. Clin Chim Acta. 2023;548:117519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 28. | Rahman A, Jahangir C, Lynch SM, Alattar N, Aura C, Russell N, Lanigan F, Gallagher WM. Advances in tissue-based imaging: impact on oncology research and clinical practice. Expert Rev Mol Diagn. 2020;20:1027-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Corti C, Cobanaj M, Dee EC, Criscitiello C, Tolaney SM, Celi LA, Curigliano G. Artificial intelligence in cancer research and precision medicine: Applications, limitations and priorities to drive transformation in the delivery of equitable and unbiased care. Cancer Treat Rev. 2023;112:102498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 30. | Huang Z, Shao W, Han Z, Alkashash AM, De la Sancha C, Parwani AV, Nitta H, Hou Y, Wang T, Salama P, Rizkalla M, Zhang J, Huang K, Li Z. Artificial intelligence reveals features associated with breast cancer neoadjuvant chemotherapy responses from multi-stain histopathologic images. NPJ Precis Oncol. 2023;7:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 45] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 31. | Chan HP, Samala RK, Hadjiiski LM, Zhou C. Deep Learning in Medical Image Analysis. Adv Exp Med Biol. 2020;1213:3-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 329] [Article Influence: 65.8] [Reference Citation Analysis (0)] |
| 32. | Bergquist M, Rolandsson B, Gryska E, Laesser M, Hoefling N, Heckemann R, Schneiderman JF, Björkman-Burtscher IM. Trust and stakeholder perspectives on the implementation of AI tools in clinical radiology. Eur Radiol. 2024;34:338-347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 33. | Kim I, Kang K, Song Y, Kim TJ. Application of Artificial Intelligence in Pathology: Trends and Challenges. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 34. | Cheng JY, Abel JT, Balis UGJ, McClintock DS, Pantanowitz L. Challenges in the Development, Deployment, and Regulation of Artificial Intelligence in Anatomic Pathology. Am J Pathol. 2021;191:1684-1692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 35. | Hanna MG, Ardon O, Reuter VE, Sirintrapun SJ, England C, Klimstra DS, Hameed MR. Integrating digital pathology into clinical practice. Mod Pathol. 2022;35:152-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 64] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 36. | Kelleher M, Colling R, Browning L, Roskell D, Roberts-Gant S, Shah KA, Hemsworth H, White K, Rees G, Dolton M, Soares MF, Verrill C. Department Wide Validation in Digital Pathology-Experience from an Academic Teaching Hospital Using the UK Royal College of Pathologists' Guidance. Diagnostics (Basel). 2023;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 37. | Ardon O, Klein E, Manzo A, Corsale L, England C, Mazzella A, Geneslaw L, Philip J, Ntiamoah P, Wright J, Sirintrapun SJ, Lin O, Elenitoba-Johnson K, Reuter VE, Hameed MR, Hanna MG. Digital pathology operations at a tertiary cancer center: Infrastructure requirements and operational cost. J Pathol Inform. 2023;14:100318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 38. | Codipilly DC, Faghani S, Hagan C, Lewis J, Erickson BJ, Iyer PG. The Evolving Role of Artificial Intelligence in Gastrointestinal Histopathology: An Update. Clin Gastroenterol Hepatol. 2024;22:1170-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 39. | Lujan GM, Savage J, Shana'ah A, Yearsley M, Thomas D, Allenby P, Otero J, Limbach AL, Cui X, Scarl RT, Hardy T, Sheldon J, Plaza JA, Whitaker B, Frankel W, Parwani AV, Li Z. Digital Pathology Initiatives and Experience of a Large Academic Institution During the Coronavirus Disease 2019 (COVID-19) Pandemic. Arch Pathol Lab Med. 2021;145:1051-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 40. | Shafi S, Parwani AV. Artificial intelligence in diagnostic pathology. Diagn Pathol. 2023;18:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 80] [Reference Citation Analysis (0)] |
| 41. | Janowczyk A, Zlobec I, Walker C, Berezowska S, Huschauer V, Tinguely M, Kupferschmid J, Mallet T, Merkler D, Kreutzfeldt M, Gasic R, Rau TT, Mazzucchelli L, Eyberg I, Cathomas G, Mertz KD, Koelzer VH, Soldini D, Jochum W, Rössle M, Henkel M, Grobholz R; Swiss Digital Pathology Consortium. Swiss digital pathology recommendations: results from a Delphi process conducted by the Swiss Digital Pathology Consortium of the Swiss Society of Pathology. Virchows Arch. 2024;485:13-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 42. | Boyce BF. An update on the validation of whole slide imaging systems following FDA approval of a system for a routine pathology diagnostic service in the United States. Biotech Histochem. 2017;92:381-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 43. | Rakha EA, Vougas K, Tan PH. Digital Technology in Diagnostic Breast Pathology and Immunohistochemistry. Pathobiology. 2022;89:334-342. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 44. | Ibrahim A, Gamble P, Jaroensri R, Abdelsamea MM, Mermel CH, Chen PC, Rakha EA. Artificial intelligence in digital breast pathology: Techniques and applications. Breast. 2020;49:267-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 104] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 45. | Baydoun A, Jia AY, Zaorsky NG, Kashani R, Rao S, Shoag JE, Vince RA Jr, Bittencourt LK, Zuhour R, Price AT, Arsenault TH, Spratt DE. Artificial intelligence applications in prostate cancer. Prostate Cancer Prostatic Dis. 2024;27:37-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 28] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 46. | Stenzinger A, Alber M, Allgäuer M, Jurmeister P, Bockmayr M, Budczies J, Lennerz J, Eschrich J, Kazdal D, Schirmacher P, Wagner AH, Tacke F, Capper D, Müller KR, Klauschen F. Artificial intelligence and pathology: From principles to practice and future applications in histomorphology and molecular profiling. Semin Cancer Biol. 2022;84:129-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 47. | da Silva LM, Pereira EM, Salles PG, Godrich R, Ceballos R, Kunz JD, Casson A, Viret J, Chandarlapaty S, Ferreira CG, Ferrari B, Rothrock B, Raciti P, Reuter V, Dogdas B, DeMuth G, Sue J, Kanan C, Grady L, Fuchs TJ, Reis-Filho JS. Independent real-world application of a clinical-grade automated prostate cancer detection system. J Pathol. 2021;254:147-158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 94] [Reference Citation Analysis (0)] |
| 48. | Grobholz R, Janowczyk A, Frei AL, Kreutzfeldt M, Koelzer VH, Zlobec I. National digital pathology projects in Switzerland: A 2023 update. Pathologie (Heidelb). 2023;44:225-228. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 49. | McGenity C, Randell R, Bellamy C, Burt A, Cratchley A, Goldin R, Hubscher SG, Neil DAH, Quaglia A, Tiniakos D, Wyatt J, Treanor D. Survey of liver pathologists to assess attitudes towards digital pathology and artificial intelligence. J Clin Pathol. 2023;77:27-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 50. | Huang B, Tian S, Zhan N, Ma J, Huang Z, Zhang C, Zhang H, Ming F, Liao F, Ji M, Zhang J, Liu Y, He P, Deng B, Hu J, Dong W. Accurate diagnosis and prognosis prediction of gastric cancer using deep learning on digital pathological images: A retrospective multicentre study. EBioMedicine. 2021;73:103631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 51. | Iqbal MJ, Javed Z, Sadia H, Qureshi IA, Irshad A, Ahmed R, Malik K, Raza S, Abbas A, Pezzani R, Sharifi-Rad J. Clinical applications of artificial intelligence and machine learning in cancer diagnosis: looking into the future. Cancer Cell Int. 2021;21:270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 98] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 52. | Berbís MA, Aneiros-Fernández J, Mendoza Olivares FJ, Nava E, Luna A. Role of artificial intelligence in multidisciplinary imaging diagnosis of gastrointestinal diseases. World J Gastroenterol. 2021;27:4395-4412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (71)] |
| 53. | Lino-Silva LS, Xinaxtle DL. Artificial intelligence technology applications in the pathologic diagnosis of the gastrointestinal tract. Future Oncol. 2020;16:2845-2851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 54. | Majidova K, Handfield J, Kafi K, Martin RD, Kubinski R. Role of Digital Health and Artificial Intelligence in Inflammatory Bowel Disease: A Scoping Review. Genes (Basel). 2021;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 55. | Robertson AR, Segui S, Wenzek H, Koulaouzidis A. Artificial intelligence for the detection of polyps or cancer with colon capsule endoscopy. Ther Adv Gastrointest Endosc. 2021;14:26317745211020277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 56. | Antonelli G, Gkolfakis P, Tziatzios G, Papanikolaou IS, Triantafyllou K, Hassan C. Artificial intelligence-aided colonoscopy: Recent developments and future perspectives. World J Gastroenterol. 2020;26:7436-7443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 57. | Parasher G, Wong M, Rawat M. Evolving role of artificial intelligence in gastrointestinal endoscopy. World J Gastroenterol. 2020;26:7287-7298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 58. | Correia FP, Lourenço LC. Artificial intelligence application in diagnostic gastrointestinal endoscopy - Deus ex machina? World J Gastroenterol. 2021;27:5351-5361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (2)] |
| 59. | Visaggi P, de Bortoli N, Barberio B, Savarino V, Oleas R, Rosi EM, Marchi S, Ribolsi M, Savarino E. Artificial Intelligence in the Diagnosis of Upper Gastrointestinal Diseases. J Clin Gastroenterol. 2022;56:23-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 60. | Suzuki H, Yoshitaka T, Yoshio T, Tada T. Artificial intelligence for cancer detection of the upper gastrointestinal tract. Dig Endosc. 2021;33:254-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 61. | Korbar B, Olofson AM, Miraflor AP, Nicka CM, Suriawinata MA, Torresani L, Suriawinata AA, Hassanpour S. Deep Learning for Classification of Colorectal Polyps on Whole-slide Images. J Pathol Inform. 2017;8:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 168] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 62. | Wei JW, Suriawinata AA, Vaickus LJ, Ren B, Liu X, Lisovsky M, Tomita N, Abdollahi B, Kim AS, Snover DC, Baron JA, Barry EL, Hassanpour S. Evaluation of a Deep Neural Network for Automated Classification of Colorectal Polyps on Histopathologic Slides. JAMA Netw Open. 2020;3:e203398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 63. | Zha Y, Xue C, Liu Y, Ni J, De La Fuente JM, Cui D. Artificial intelligence in theranostics of gastric cancer, a review. Med Rev (2021). 2023;3:214-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 64. | Fraggetta F, L'Imperio V, Ameisen D, Carvalho R, Leh S, Kiehl TR, Serbanescu M, Racoceanu D, Della Mea V, Polonia A, Zerbe N, Eloy C. Best Practice Recommendations for the Implementation of a Digital Pathology Workflow in the Anatomic Pathology Laboratory by the European Society of Digital and Integrative Pathology (ESDIP). Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 65. | Calderaro J, Kather JN. Artificial intelligence-based pathology for gastrointestinal and hepatobiliary cancers. Gut. 2021;70:1183-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 66. | Kather JN, Calderaro J. Development of AI-based pathology biomarkers in gastrointestinal and liver cancer. Nat Rev Gastroenterol Hepatol. 2020;17:591-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 67. | Murchan P, Ó'Brien C, O'Connell S, McNevin CS, Baird AM, Sheils O, Ó Broin P, Finn SP. Deep Learning of Histopathological Features for the Prediction of Tumour Molecular Genetics. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 68. | Acs B, Rantalainen M, Hartman J. Artificial intelligence as the next step towards precision pathology. J Intern Med. 2020;288:62-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 224] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 69. | Wang Y, Hu C, Kwok T, Bain CA, Xue X, Gasser RB, Webb GI, Boussioutas A, Shen X, Daly RJ, Song J. DEMoS: a deep learning-based ensemble approach for predicting the molecular subtypes of gastric adenocarcinomas from histopathological images. Bioinformatics. 2022;38:4206-4213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 70. | Echle A, Grabsch HI, Quirke P, van den Brandt PA, West NP, Hutchins GGA, Heij LR, Tan X, Richman SD, Krause J, Alwers E, Jenniskens J, Offermans K, Gray R, Brenner H, Chang-Claude J, Trautwein C, Pearson AT, Boor P, Luedde T, Gaisa NT, Hoffmeister M, Kather JN. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology. 2020;159:1406-1416.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 220] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 71. | Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24:1248-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 589] [Article Influence: 84.1] [Reference Citation Analysis (0)] |
| 72. | Liao H, Long Y, Han R, Wang W, Xu L, Liao M, Zhang Z, Wu Z, Shang X, Li X, Peng J, Yuan K, Zeng Y. Deep learning-based classification and mutation prediction from histopathological images of hepatocellular carcinoma. Clin Transl Med. 2020;10:e102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 73. | Schmauch B, Romagnoni A, Pronier E, Saillard C, Maillé P, Calderaro J, Kamoun A, Sefta M, Toldo S, Zaslavskiy M, Clozel T, Moarii M, Courtiol P, Wainrib G. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat Commun. 2020;11:3877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 263] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 74. | Huang Z, Johnson TS, Han Z, Helm B, Cao S, Zhang C, Salama P, Rizkalla M, Yu CY, Cheng J, Xiang S, Zhan X, Zhang J, Huang K. Deep learning-based cancer survival prognosis from RNA-seq data: approaches and evaluations. BMC Med Genomics. 2020;13:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 75. | Pandey D, Onkara Perumal P. A scoping review on deep learning for next-generation RNA-Seq. data analysis. Funct Integr Genomics. 2023;23:134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 76. | Wei JW, Wei JW, Jackson CR, Ren B, Suriawinata AA, Hassanpour S. Automated Detection of Celiac Disease on Duodenal Biopsy Slides: A Deep Learning Approach. J Pathol Inform. 2019;10:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 77. | Tomita N, Abdollahi B, Wei J, Ren B, Suriawinata A, Hassanpour S. Attention-Based Deep Neural Networks for Detection of Cancerous and Precancerous Esophagus Tissue on Histopathological Slides. JAMA Netw Open. 2019;2:e1914645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 78. | Iizuka O, Kanavati F, Kato K, Rambeau M, Arihiro K, Tsuneki M. Deep Learning Models for Histopathological Classification of Gastric and Colonic Epithelial Tumours. Sci Rep. 2020;10:1504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 215] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 79. | Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph. 2017;61:2-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 172] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 80. | Xiao Y, Wang S, Ling R, Song Y. Application of artificial neural network algorithm in pathological diagnosis and prognosis prediction of digestive tract malignant tumors. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2023;52:243-248. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 81. | Noorbakhsh J, Farahmand S, Foroughi Pour A, Namburi S, Caruana D, Rimm D, Soltanieh-Ha M, Zarringhalam K, Chuang JH. Deep learning-based cross-classifications reveal conserved spatial behaviors within tumor histological images. Nat Commun. 2020;11:6367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 114] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 82. | Kuntz S, Krieghoff-Henning E, Kather JN, Jutzi T, Höhn J, Kiehl L, Hekler A, Alwers E, von Kalle C, Fröhling S, Utikal JS, Brenner H, Hoffmeister M, Brinker TJ. Gastrointestinal cancer classification and prognostication from histology using deep learning: Systematic review. Eur J Cancer. 2021;155:200-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 87] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 83. | Huang G, Wang C, Fu X. Bidirectional deep neural networks to integrate RNA and DNA data for predicting outcome for patients with hepatocellular carcinoma. Future Oncol. 2021;17:4481-4495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 84. | Qi L, Liang JY, Li ZW, Xi SY, Lai YN, Gao F, Zhang XR, Wang DS, Hu MT, Cao Y, Xu LJ, Chan RCK, Xing BC, Wang X, Li YH. Deep learning-derived spatial organization features on histology images predicts prognosis in colorectal liver metastasis patients after hepatectomy. iScience. 2023;26:107702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 85. | Wang JG. Application and future perspectives of gastric cancer technology based on artificial intelligence. Tzu Chi Med J. 2023;35:148-151. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 86. | Caputo A, L'Imperio V, Merolla F, Girolami I, Leoni E, Della Mea V, Pagni F, Fraggetta F. The slow-paced digital evolution of pathology: lights and shadows from a multifaceted board. Pathologica. 2023;115:127-136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 87. | Tizhoosh HR, Pantanowitz L. Artificial Intelligence and Digital Pathology: Challenges and Opportunities. J Pathol Inform. 2018;9:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 260] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 88. | Browning L, Jesus C, Malacrino S, Guan Y, White K, Puddle A, Alham NK, Haghighat M, Colling R, Birks J, Rittscher J, Verrill C. Artificial Intelligence-Based Quality Assessment of Histopathology Whole-Slide Images within a Clinical Workflow: Assessment of 'PathProfiler' in a Diagnostic Pathology Setting. Diagnostics (Basel). 2024;14. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 89. | Yoshida H, Kiyuna T. Requirements for implementation of artificial intelligence in the practice of gastrointestinal pathology. World J Gastroenterol. 2021;27:2818-2833. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 90. | Wong ANN, He Z, Leung KL, To CCK, Wong CY, Wong SCC, Yoo JS, Chan CKR, Chan AZ, Lacambra MD, Yeung MHY. Current Developments of Artificial Intelligence in Digital Pathology and Its Future Clinical Applications in Gastrointestinal Cancers. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 91. | Giansanti D. The Regulation of Artificial Intelligence in Digital Radiology in the Scientific Literature: A Narrative Review of Reviews. Healthcare (Basel). 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 92. | Yao L, Lu Z, Yang G, Zhou W, Xu Y, Guo M, Huang X, He C, Zhou R, Deng Y, Wu H, Chen B, Gong R, Zhang L, Zhang M, Gong W, Yu H. Development and validation of an artificial intelligence-based system for predicting colorectal cancer invasion depth using multi-modal data. Dig Endosc. 2023;35:625-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 93. | Abels E, Pantanowitz L. Current State of the Regulatory Trajectory for Whole Slide Imaging Devices in the USA. J Pathol Inform. 2017;8:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 94. | García-Rojo M, De Mena D, Muriel-Cueto P, Atienza-Cuevas L, Domínguez-Gómez M, Bueno G. New European Union Regulations Related to Whole Slide Image Scanners and Image Analysis Software. J Pathol Inform. 2019;10:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |