Published online Dec 28, 2022. doi: 10.35712/aig.v3.i5.117
Peer-review started: October 9, 2022
First decision: October 29, 2022
Revised: November 21, 2022
Accepted: December 21, 2022
Article in press: December 21, 2022
Published online: December 28, 2022
Processing time: 79 Days and 8.7 Hours
Artificial intelligence (AI) is a complex concept, broadly defined in medicine as the development of computer systems to perform tasks that require human intelligence. It has the capacity to revolutionize medicine by increasing efficiency, expediting data and image analysis and identifying patterns, trends and associations in large datasets. Within gastroenterology, recent research efforts have focused on using AI in esophagogastroduodenoscopy, wireless capsule endoscopy (WCE) and colonoscopy to assist in diagnosis, disease monitoring, lesion detection and therapeutic intervention. The main objective of this narrative review is to provide a comprehensive overview of the research being performed within gastroenterology on AI in esophagogastroduodenoscopy, WCE and colonoscopy.
Core Tip: Artificial intelligence (AI) is a complex concept that has the capacity to revolutionize medicine. Within gastroenterology, recent research efforts have focused on using AI in esophagogastroduodenoscopy, wireless capsule endoscopy (WCE) and colonoscopy to assist in diagnosis, disease monitoring, lesion detection and therapeutic intervention. This narrative review provides a comprehensive overview of the research being performed within gastroenterology on AI in esophagogastroduodenoscopy, WCE and colonoscopy.
- Citation: Galati JS, Duve RJ, O'Mara M, Gross SA. Artificial intelligence in gastroenterology: A narrative review. Artif Intell Gastroenterol 2022; 3(5): 117-141
- URL: https://www.wjgnet.com/2644-3236/full/v3/i5/117.htm
- DOI: https://dx.doi.org/10.35712/aig.v3.i5.117
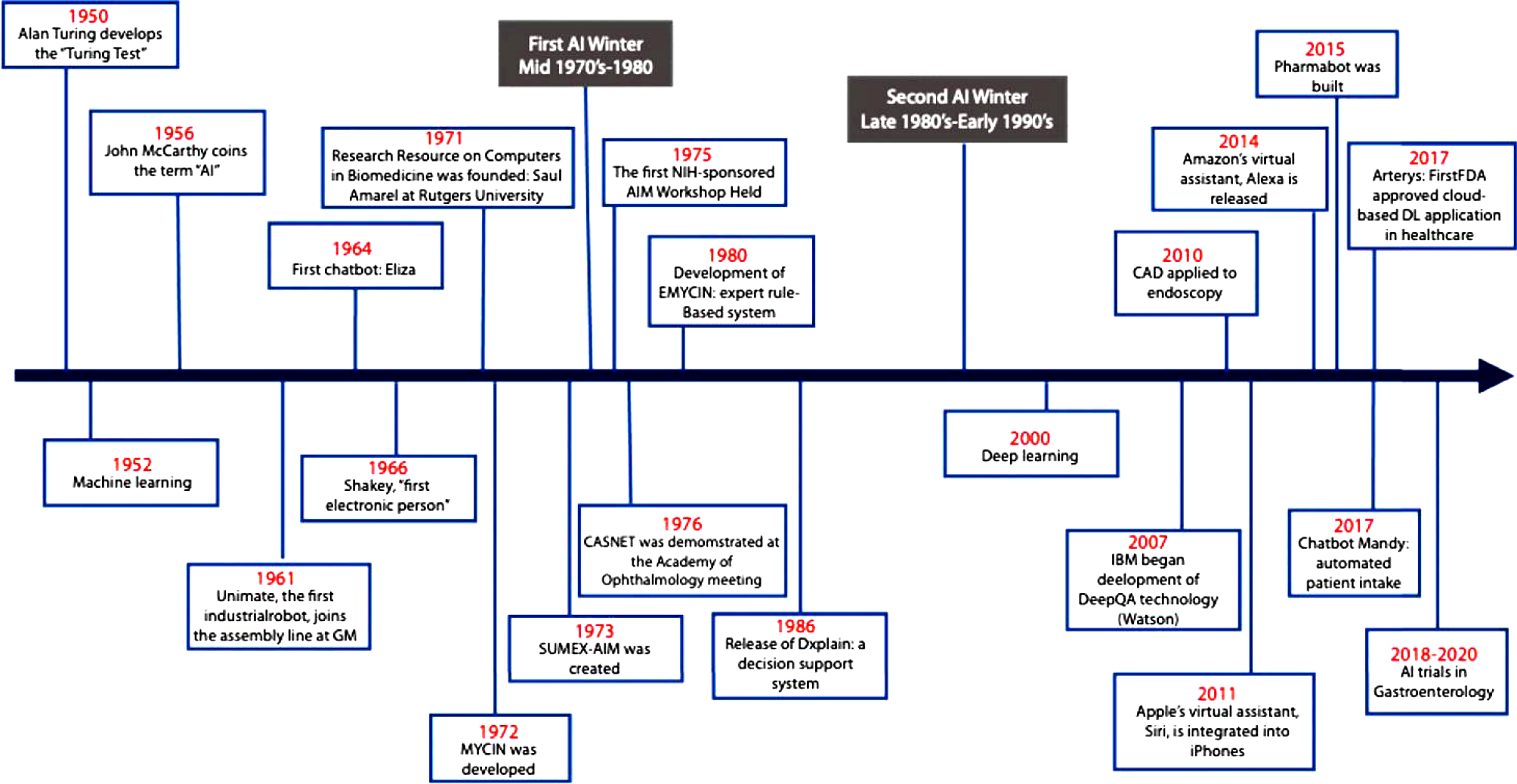
Artificial intelligence (AI) is a complex concept, broadly defined in medicine as the development of computer systems to perform tasks that require human intelligence[1]. Since its inception in the 1950s, the field of AI has grown considerably (Figure 1)[2]. Often AI is accompanied by the terms machine learning (ML) and deep learning (DL), techniques used within the field of AI to develop systems that can learn and adapt without explicit instructions. Machine learning uses self-learning algorithms that derive knowledge from data to predict outcomes[1]. There are two main categories within ML: Supervised and unsupervised learning. In supervised learning, the AI is trained on a dataset in which human intervention has previously assigned a hierarchy of features which allows the algorithm to understand differences between data inputs and classify or predict outcomes[3]. In unsupervised learning, the system is provided a dataset that has not been categorized by human intervention. The algorithm then analyzes the data with the goal of identifying labels or patterns[3].
Deep learning is a subfield of ML that utilizes artificial neural networks (ANN) to analyze data. In DL, the system is able to analyze raw data and determine features that distinguish between data inputs. ANN systems are composed of interconnected nodes in a layered structure similar to how neurons are organized in the human brain. The weight of the connections between each node influences how the system can recognize, classify, and describe objects within data[3,4]. ANNs with multiple layers of nodes are classified as deep neural networks which form the backbone of deep learning.
Artificial intelligence has the capacity to revolutionize medicine. It can be used to increase efficiency by aiding in appointment scheduling, reviewing insurance eligibility, or tracking patient history. AI can also expedite data and image analysis and detect patterns, trends and associations[5]. Within gastroenterology, AI’s prominence stems from its utility in image analysis[5,6]. Many gastrointestinal diseases rely on endoscopic evaluation for diagnosis, disease monitoring, lesion detection and therapeutic intervention. However, endoscopic evaluation is heavily operator dependent and thus subject to operator bias and human error. As such, recent efforts have focused on using AI in esophagogastroduodenoscopy, wireless capsule endoscopy (WCE) and colonoscopy to mitigate these issues, serving as an additional objective observer of the intestinal tract. The main objective of this narrative review is to provide a comprehensive overview of the research being performed within gastroenterology on artificial intelligence in esophagogastroduodenoscopy, WCE and colonoscopy. While other narrative reviews have been published regarding the use of artificial intelligence in esophagogastroduodenoscopy, WCE and colonoscopy, this narrative review goes a step further by providing a granular and more technical assessment of the literature. As such, this narrative review is intended for medical providers and researchers who are familiar with the use of artificial intelligence in esophagogastroduodenoscopy, WCE and colonoscopy and are interested in obtaining an in-depth review in a specific area.
Electronic databases Embase, Ovid Medicine, and PubMed were searched from inception to September 2022 using multiple search queries. Combinations of the terms “artificial intelligence”, “AI”, “computer aided”, “computer aided detection”, “CADe”, “convolutional neural network”, “deep learning”, “DCNN”, “machine learning”, “colonoscopy”, “endoscopy”, “wireless capsule endoscopy”, “capsule endoscopy”, “WCE”, “esophageal cancer”, esophageal adenocarcinoma”, “esophageal squamous cell carcinoma”, “gastric cancer”, “gastric neoplasia”, “gastric lesions”, “Barrett’s esophagus”, “celiac disease”, “Helicobacter pylori”, “Helicobacter pylori infection”, “H pylori”, “H pylori infection”, “gastric ulcers”, “duodenal ulcers”, “inflammatory bowel disease”, “IBD”, “ulcerative colitis”, “Crohn’s disease”, “parasitic infections”, “hookworms”, “bleeding”, “gastrointestinal bleeding”, “vascular lesions”, “angioectasias”, “polyp”, “polyp detection”, “tumor”, “gastrointestinal tumor”, “small bowel tumor”, “bowel preparation”, “Boston bowel preparation scale”, “BBPS”, “adenoma”, “adenoma detection”, “adenoma detection rate”, “sessile serrated lesion”, and “sessile serrated lesion rate” were used. We subsequently narrowed the results to clinical trials in human published within the last 10 years.
Barrett’s esophagus (BE) is a premalignant condition associated with esophageal adenocarcinoma (EAC)[7-9]. It is caused by chronic inflammation and tissue injury of the lower esophagus as a result of gastric reflux[7-9]. Early detection and diagnosis can prevent the progression of BE to EAC[7-9]. Patients with BE should undergo routine surveillance endoscopies to monitor for progression. However, even with surveillance, dysplastic changes can be easily missed[7]. To improve the detection of dysplastic changes in BE, researchers have focused on developing AI systems to assist with the identification of dysplasia and early neoplasia during endoscopic evaluation.
Since 2016, a group of researchers from the Netherlands have developed numerous AI systems to identify neoplastic lesions in BE[10-16]. Their first publication detailed their experience using a support vector machine (SVM), a ML method, to identify early neoplastic lesions from white light endoscopy (WLE) images[10]. Their SVM achieved a sensitivity and specificity of 83% with respect to per-image detection and sensitivity of 86% and specificity of 87% with respect to per-patient detection[10]. In their next study, the group trialed several different feature extraction and ML methods using volumetric laser endomicroscopy (VLE) images[11]. They received the best results with the feature extraction module “layering and signal decay statistics”, achieving high sensitivity (90%) and specificity (93%) with area under the curve (AUC) 0.95 for neoplastic lesion detection[11]. Following this, they conducted a second studying again using ML in VLE to identify neoplastic lesions in BE, however, they used a multiframe analysis approach, including frames neighboring the region of interest in the analysis[12]. With this approach, they found that multiframe analysis resulted in a significantly higher median AUC when compared to single frame analysis (0.91 vs 0.83; P < 0.001)[12]. Continuing to use ML methods, the group published their finding from the ARGOS project – a consortium of three international tertiary referral centers for Barrett’s neoplasia[13]. In this study, de Groof et al[13] created a computer-aided detection (CADe) system that used SVM to classify images. The group tested the CADe with 60 images – 40 images from patients with a neoplastic lesion, 20 images from patients with non-dysplastic Barrett’s esophagus. The CADe achieved an AUC of 0.92 and a sensitivity, specificity and accuracy of 95%, 85% and 92% respectively for detecting neoplastic lesions[13].
Following their successes creating ML systems for neoplastic lesion detection, the group of researchers from the Netherlands shifted their focus to DL methods. In their first foray into DL, they developed a hybrid CADe system using architecture from ResNet and U-Net models. The CADe was trained with 494364 labeled endoscopic images and subsequently refined with a data set comprised of 1247 WLE images. It was finally tested on a set of 297 images (129 images with early neoplasia, 168 with non-dysplastic BE) where the hybrid CADe system attained a sensitivity of 87.6%, specificity of 88.6% and accuracy of 88.2% for identifying early neoplasia[14]. The system was also tested in two external validation sets where it achieved similar results. A secondary outcome of the study was to see if within the images classified as having neoplasia if the CADe could delineate the neoplasia and recommend a site for biopsy. The ground truth was determined by expert endoscopists. In two external data sets (external validation data set 4 and 5), the CADe identified the optimal biopsy site in 97.2% of cases and 91.9% of cases respectively[14]. Using a similar hybrid CADe, the group performed a pilot study testing the CADe during live endoscopic procedures[15]. Overall, the CADe achieved a sensitivity of 75.8%, specificity of 86.5% and accuracy of 84% in per-image analyses[15]. Their most recent study again used their hybrid ResNet and U-Net CADe to identify neoplastic lesions in narrow-band imaging (NBI)[16]. With respect to NBI images, the CADe was found to have sensitivity of 88% (95%CI 86%-94%), specificity of 78% (95%CI 72%-84%), and accuracy of 84% (95%CI 81%-88%) for identifying BE neoplasia[16]. In per frame and per video analyses, the CADe achieved sensitivities of 75% and 85%, specificities of 90% and 83% and accuracies of 85% and 83% respectively[16].
Outside of this group from the Netherlands, several other researchers have created DL systems for the detection of BE neoplasia[17-21]. Hong et al[17] created a CNN that could distinguish between intestinal metaplasia, gastric metaplasia and neoplasia from images obtained by endomicroscopy in patients with Barrett’s esophagus with accuracy of 80.8%. Ebigbo et al[18] created a DL-CADe capable of detecting BE neoplasia with sensitivity 83.7%, specificity of 100.0% and accuracy of 89.9%. Two other groups achieved similar results to Ebigbo et al[18]: Hashimoto et al’s CNN detected early neoplasia with sensitivity of 96.4%, specificity of 94.2%, and accuracy of 95.4% and Hussein et al’s CNN detected early neoplasia with sensitivity 91%, specificity 79%, area under the receiver operating characteristic (AUROC) of 93%[19,20]. An overview of these studies is provided in Table 1.
| Ref. | Country | Study design | AI Classifier | Lesions | Training dataset | Test dataset | Sensitivity (%) | Specificity (%) | Accuracy (%) | AUROC |
| Swager et al[11], 2017 | Netherlands | Retrospective | ML2 methods | NPL | - | 60 VLE images | 90 | 93 | - | 0.95 |
| van der Sommen et al[10], 2016 | Netherlands | Retrospective | SVM | NPL | - | 100 WLE images | 83 | 83 | - | - |
| Hong et al[17], 2017 | South Korea | Retrospective | CNN | NPL, IM, GM | 236 endomicroscopy images | 26 endomicroscopy images | - | - | 80.77 | - |
| de Groof et al[13], 2019 | Netherlands, Germany, Belgium | Prospective | SVM | NPL | - | 60 WLE images | 95 | 85 | 91.7 | 0.92 |
| Ebigbo et al[21], 2019 | Germany, Brazil | Retrospective | CNN | EAC | Augsburg dataset: 148 WLE images and NBI; MICCAI dataset: 100 WLE images | 97; 94a; 92 | 88; 80a; 100 | - | - | |
| Ghatwary et al[24], 2019 | England, Egypt | Retrospective | Multiple CNNs | EAC | Images from 21 patients | Images from 9 patients | 96 | 92 | - | - |
| de Groof et al[14], 2020 | Netherlands, France, Sweden, Germany, Belgium, Australia | Ambispective | CNN | NPL | Dataset 1: 494364 images; Dataset 2:1; 247 images; Dataset 3: 297 images | Dataset 3: 297 images; Dataset 4: 80 images; Dataset 5: 80 images | 90b | 87.5b | 88.8b | - |
| de Groof et al[15], 2020 | Netherlands, Belgium | Prospective | CNN | NPL | 495611 images | 20 patients; 144 WLE images | 75.8 | 86.5 | 84 | - |
| Ebigbo et al[18], 2020 | Germany, Brazil | Prospective | CNN | EAC | 129 images | 62 images | 83.7 | 100 | 89.9 | - |
| Hashimoto et al[19], 2020 | United States | Retrospective | CNN | NPL | 1374 images | 458 images | 96.4 | 94.2 | 95.4 | - |
| Struyvenberg et al[12], 2020 | Netherlands | Prospective | ML2 methods | NPL | - | 3060 VLE frames | - | - | - | 0.91 |
| Iwagami et al[25], 2021 | Japan | Retrospective | CNN | EJC | 3443 images | 232 images | 94 | 42 | 66 | - |
| Struyvenberg et al[16], 2021 | Netherlands, Sweden, Belgium | Retrospective | CNN | NPL | 495611 images | 157 NBI zoom videos; 30021 frames | 851; 75 | 831; 90 | 831; 85 | - |
| Hussein et al[20], 2022 | England, Spain, Belgium, Austria | Prospective | CNN | DPL | 148936 frames | 264 iscan-1 images | 91 | 79 | - | 0.93 |
In addition to neoplasia detection, some groups started to use AI to grade BE and predict submucosal invasion of lesions. Ali et al[22] recently published the results from a pilot study using a DL system to quantitatively assess BE area (BEA), circumference and maximal length (C&M). They tested their DL system on 3D printed phantom esophagus models with different BE patterns and 194 videos from 131 patients with BE. In the phantom esophagus models, the DL system achieved an accuracy of 98.4% for BEA and 97.2% for C&M[22]. In the patient videos, the DL system differed from expert endoscopists by 8% and 7% for C&M respectively[22]. Ebigbo et al[23], building upon their earlier success using a DL CADe to detect neoplasia, performed a pilot study using a 101-layer CNN to differentiate T1a (mucosal) and T1b (submucosal) BE related cancers. Using 230 WLE images obtained from three tertiary care centers in Germany, their CNN was capable of discerning T1a lesions from T1b lesions with sensitivity, specificity and accuracy of 77%, 64% and 71% respectively, comparable to the expert endoscopists enrolled in the study[23].
Despite BE’s potential progression to EAC if left unmanaged, few studies have explicitly looked at using AI to detect EAC. Ghatwary et al[24] tested several DL models on 100 WLE images (50 featuring EAC, 50 featuring normal mucosa) to determine which was best at identifying EAC. They found that the Single-Shot Multibox Detector (SSD) method achieved the best results, attaining a sensitivity of 96% and specificity of 92%[24]. In 2021, Iwagami et al[25] focused on developing an AI system to identify esophagogastric junctional adenocarcinomas. They used SSD for their CNN, achieving a sensitivity, specificity and accuracy of 94%, 42% and 66% for detecting esophagogastric junctional adenocarcinomas. Their CNN performed similarly to endoscopists enrolled in the study (sensitivity 88%, specificity 43%, accuracy 66%)[25].
Esophageal squamous cell carcinoma (ESCC) is the most common histologic type of esophageal cancer in the world[26]. While certain imaging modalities such as Lugol’s chromoendoscopy and confocal microendoscopy are effective at improving the accuracy, sensitivity and specificity of targeted biopsies, they are expensive and not universally available[27]. In recent years, efforts have focused on developing AI systems to support lower cost imaging modalities in order to improve their ability to detect ESCC.
Shin et al[27] and Quang et al[28] created ML algorithms which they tested on high-resolution microendoscope images, obtaining comparable sensitivities for the detection of ESCC (98% and 95% respectively). Following these studies, several groups created DL systems to detect ESCC[29-38]. In Cai et al’s study, their deep neural network-CADe was tested on 187 images obtained from WLE. The system obtained good sensitivity (97.8%), specificity (85.4%) and accuracy (91.4%) for identifying ESCC[29]. Similar findings occurred in three separate studies that used deep convolutional neural networks (DCNNs) to detect ESCC in WLE[30-32]. Using NBI, Guo et al[33] created a CADe that achieved high sensitivity (98.0%), specificity (95.0%) and an AUC of 0.99 for detecting ESCC in still images. Similar results were obtained in Li et al’s study[35]. For detecting ESCC in NBI video clips, Fukuda et al[34] obtained different results, finding similar sensitivity (91%) to Guo et al[33] however substantially lower specificity (51%). Three studies compared a DL-CADe with WLE to DL-CADe with NBI for the detection of ESCC[32,35,36]. The results from these three studies were quite discordant and as such a statement regarding whether a DL-CADe with WLE or DL-CADe with NBI is better for the detection of ESCC cannot be made at this time.
Interestingly, several studies used DL algorithms to assess ESCC invasion depth[39,40]. Everson et al[39] and Zhao et al[40] created CNNs to detect intrapapillary capillary loops, a feature of ESCC that correlates with invasion depth, in images obtained from magnification endoscopy with NBI. They achieved similar findings with Everson et al’s CNN achieving an accuracy of 93.7% and Zhao et al’s achieving an accuracy of 89.2%[39,40]. Using DL, two groups created DCNNs to directly detect ESCC invasion depth[41-43]. One group from Osaka International Cancer Institute conducted two studies using SSD to create their DCNNs[41,42]. The DCNNs were made to classify images as EP-SM1 or EP-SM2-3 as this distinction in ESCC bares clinical significance. The studies (Nakagawa et al[41] and Shimamoto et al[42]) attained similar accuracies and specificities, however had substantially different sensitivities (90.1% vs 50% and 71%)[41,42]. The third study, Tokai et al[43], used SSD as well for their DCNN and also programed the DCNN to classify images as EP-SM1 or EP-SM2-3. Their observed sensitivity, specificity and accuracy were lower than those found by Nakagawa et al[41] (84.1%, 73.3% and 80.9% respectively).
Gastric cancer is the third leading cause of cancer-related mortality in the world[44,45]. Early detection of precancerous lesions or early gastric cancer with endoscopy can prevent progression to advanced disease[46]. However, a substantial number of upper gastrointestinal cancers are missed placing patients at risk for interval development[45]. To mitigate this risk, AI systems are being develop to assist with lesion detection.
In 2013, Miyaki et al[47] used a bag-of-features framework with densely sampled scale-invariant feature transform descriptors to classify still images obtained from magnifying endoscopy with flexible spectral imaging color enhancement as having or not having gastric cancer. Their system, a rudimentary version of ML, obtained good sensitivity (84.8%), specificity (87.0%) and accuracy (85.9%) for identifying gastric cancer[47]. Using SVM, Kanesaka et al[48] found higher sensitivity (96.7%), specificity (95%) and accuracy (96.3%).
Following these successes, several groups began using CNNs for the identification of gastric cancer[44-46,49-59]. In 2018, Hirasawa et al[44] published one of the first papers to use a CNN (SSD) to detect gastric cancer. In a test set of 2296 images, the CNN had a sensitivity of 92.2% for identifying gastric cancer lesions[44]. In a larger study, Tang et al[49] created a DCNN to detect gastric cancer in a test set of 9417 images and 26 endoscopy videos. With respect to their test set, the DCNN performed well, achieving a sensitivity of 95.5% (95%CI 94.8%–96.1%), specificity of 81.7% (95%CI 80.7%–82.8%), accuracy of 87.8% (95%CI 87.1%–88.5%) and AUC 0.94[49]. The DCNN continued to perform well in external validation sets, achieving sensitivity of 85.9%-92.1%, specificity of 84.4%-90.3%, accuracy of 85.1%-91.2% and AUC 0.89-0.93[49]. Compared to expert endoscopists, the DCNN attained higher sensitivity, specificity and accuracy. In the video set, the DCNN achieved a sensitivity of 88.5% (95%CI 71.0%-96.0%)[49]. Several studies using DCNN to detect gastric cancer in endoscopy images obtained similar sensitivities, specificities and accuracies to Tang et al[49]. While one study reported a sensitivity of 58.4% for detecting gastric cancer, the sensitivity for the study’s 67 endoscopists was 31.9%[55].
Recently, several groups from China and Japan have published studies using CNNs with magnified endoscopy with NBI (ME-NBI) in an effort to improve early gastric cancer detection[56-59]. Using a 22-layer CNN, Horiuchi et al[56] achieved a sensitivity, specificity and accuracy of 95.4%, 71.0% and 85.3% respectively for identifying early gastric cancer from a set of 258 ME-NBI images (151 gastric cancer, 107 gastritis). The same group published a similar study the following year however using ME-NBI videos instead of still images[57]. They obtained similar results: sensitivity of 87.4% (95%CI 78.8%-92.8%), specificity of 82.8% (95%CI 73.5%-89.3%) and accuracy of 85.1% (955 CI 79.0%-89.6%)[57]. Hu et al[58] and Ueyama et al[59] in their studies using CNN to identify gastric cancer in ME-NBI achieved similar sensitivities, specificities and accuracies as Horiuchi et al[56]. An overview of these studies is provided in Table 2.
| Ref. | Country | Study design | AI classifier | Lesions | Training dataset | Test dataset | Sensitivity (%) | Specificity (%) | Accuracy (%) | AUROC |
| Miyaki et al[47], 2013 | Japan | Prospectivea | SVM | Gastric cancer | 493 FICE-derived magnifying endoscopic images | 92 FICE-derived magnifying endoscopic images | 84.8 | 97 | 85.9 | - |
| Kanesaka et al[48], 2018 | Japan, Taiwan | Retrospective | SVM | EGC | 126 M-NBI images | 81 M-NBI images | 96.7 | 95 | 96.3 | - |
| Wu et al[50], 2019 | China | Retrospective | CNN | EGC | 9151 images | 200 images | 94 | 91 | 92.5 | - |
| Cho et al[51], 2019 | South Korea | Ambispective | CNN | Advanced gastric cancer, EGC, high grade dysplasia, low grade dysplasia, non-neoplasm | 4205 WLE images | 812 WLE images; 200 WLE images | - | - | 86.6b; 76.4 | 0.877b |
| Tang et al[49], 2020 | China | Retrospective | CNN | EGC | 35823 WLE images | Internal: 9417 WLE images; External: 1514 WLE images1 | 95.51; 85.9-92.1 | 81.71; 84.4-90.3 | 87.81; 85.1-91.2 | 0.941; 0.887-0.925 |
| Namikawa et al[52], 2020 | Japan | Retrospective | CNN | Gastric cancer | 18410 images | 1459 images | 99 | 93.3 | 99 | - |
| Horiuchi et al[56], 2020 | Japan | Retrospective | CNN | EGC | 2570 M-NBI images | 258 M-NBI images | 95.4 | 71 | 85.3 | 0.852 |
| Horiuchi et al[57], 2020 | Japan | Retrospective | CNN | EGC | 2570 M-NBI images | 174 videos | 87.4 | 82.8 | 85.1 | 0.8684 |
| Guo et al[54], 2021 | China | Retrospective | CNN | Gastric cancer, erosions/ulcers, polyps, varices | 293162 WLE images | 33959 WLE images | 67.52; 85.1 | 70.92; 90.3 | - | - |
| Ikenoyama et al[55], 2021 | Japan | Retrospective | CNN | EGC | 13584 WLE and NBI images | 2940 WLE and NBI images | 58.4 | 87.3 | - | - |
| Hu et al[58], 2021 | China | Retrospective | CNN | EGC | M-NBI images from 170 patients | Internal: M-NBI from 73 patients External: M-NBI images from 52 patients | 79.23; 78.2 | 74.53; 74.1 | 773; 76.3 | 0.8083; 0.813 |
| Ueyama et al[59], 2021 | Japan | Retrospective | CNN | EGC | 5574 M-NBI images | 2300 M-NBI | 98 | 100 | 98.7 | - |
| Yuan et al[53], 2022 | China | Retrospective | CNN | EGC, advanced gastric cancer, submucosal tumor, polyp, peptic ulcer, erosion, and lesion-free gastric mucosa | 29809 WLE images | 1579 WLE images | 59.24; 100 | 99.34; 98.1 | 93.54; 98.4 | - |
Of increasing interest to researchers within this field is predicting invasion depth of gastric cancer using AI. Few studies have used CNNs to predict invasion depth[60-63]. Yoon et al[60] created a CNN to predict gastric cancer lesion depth from standard endoscopy images. The CNN achieved good sensitivity (79.2%) and specificity (77.8%) for differentiating T1a (mucosal) from T1b (submucosal) gastric cancers (AUC 0.851)[60]. Also using standard endoscopy images, Zhu et al[61] attained similar results. They trained their CNN to identify P0 (restricted to the mucosa or < 0.5 mm within the muscularis mucosae) vs P1 (≥ 0.5 mm deep into the muscularis mucosae) lesions. The CNN achieved a sensitivity of 76.6%, specificity of 95.6%, accuracy of 89.2% and AUROC 0.94 (95%CI 0.90-0.97). Cho et al[62] using DenseNet-161 as their CNN and Nagao et al[62] using ResNet50 as their CNN obtained comparable results to Zhu et al[61] for predicting gastric cancer invasion depth from endoscopy images.
Within recent years, numerous studies have been published regarding the use of AI to assist with the detection and classification of gastric lesions. Few of these studies explicitly used AI systems to detect duodenal and gastric ulcers, however they report data pertaining to ulcer detection.
Using YOLOv5, a deep learning object detection model, Ku et al[64] created a CADe system capable of detecting multiple gastric lesions with good precision (98%) and sensitivity (89%). Also using YOLO for their DCNN, Yuan et al[53] achieved an overall system accuracy of 85.7% for gastric lesion identification. With respect to peptic ulcer detection, their system achieved an accuracy of 95.4% (93.5%-97.2%), sensitivity of 86.2% (77.5%–94.8%) and specificity of 96.8% (95.1%–98.4%)[53]. Guo et al[54] used ResNet50 to construct their CADe designed to detect gastric lesions. Their CADe achieved lower sensitivity 71.4% (95%CI 69.5–73.2%) and specificity 70.9% (95%CI 70.3–71.4%) than Yuan et al’s DCNN[53], however Guo et al[54] combined erosions and ulcers into one category for analysis. With their primary outcome being classifying gastric cancers and ulcers, Namikawa et al[52] developed a CNN capable of identifying gastric ulcers with high sensitivity (93.3%; 95%CI 87.3%−97.1%) and specificity (99.0%; 95%CI 94.6%-100%).
As a risk factor for future development of gastric cancer, early detection and eradication of Helicobacter pylori (H. pylori) in infected individuals is important. Endoscopic evaluation for H. pylori is highly operator dependent[65]. Pairing artificial intelligence with endoscopy for the detection of H. pylori could possibly reduce false results.
Shichijo et al[66] used GoogLeNet, a DCNN consisting of 22 layers, to evaluate 11481 images obtained from 397 patients (72 H. pylori positive, 325 negative) for the presence or absence of H. pylori infection. GoogLeNet attained a sensitivity of 81.9% (95%CI 71.1%-90.0%), specificity of 83.4% (95%CI 78.9%-87.3%) and accuracy of 83.1% (95%CI 79.1%-86.7%) with AUROC 0.89 for detecting H. Pylori infection[66]. When compared to endoscopists enrolled in the study, the sensitivity, specificity and accuracy attained by GoogLeNet was comparable to those attained by the endoscopists[66]. This same group published a second study in 2019 again using GoogLeNet for their DCNN[67]. However, a different optimization technique was used to prepare GoogLeNet. The DCNN was tasked with classifying images as H. pylori positive, negative or eradicated. In a set of 23699 images, the DCNN attained an accuracy of 80% for H. pylori negative, 84% for H. pylori eradicated, and 48% for H. pylori positive[67]. Also using GoogLeNet, Itoh et al[68] obtained similar results to Shichijo et al’s 2017 study with respect to sensitivity (86.7%) and specificity (86.7%)[66]. Using ResNet-50 as their architectural unit for their DCNN, Zheng et al[69] were successful in classifying images as H. pylori positive or negative, achieving a sensitivity, specificity, accuracy and AUC of 81.4% (95%CI 79.8%–82.9%), 90.1% (95%CI 88.4%–91.7%), 84.5% (95%CI 83.3%–85.7%) and 0.93 (95%CI 0.92–0.94) respectively.
Taking a different approach, Yasuda et al[70] used linked color imaging (LCI) with SVM to identify H. pylori infection. The LCI images were classified into high-hue and low-hue images based on redness and classified by SVM as H. pylori positive or negative. This method attained a sensitivity, specificity and accuracy of 90.4%, 85.7% and 87.6% respectively[70]. Combining LCI with a deep learning CADe system, Nakashima et al[71] achieved a sensitivity, specificity and accuracy of 92.5%, 80.0%, 84.2% for identifying H. pylori negative images, 62.5%, 92.5%, 82.5% for H pylori positive images, 65%, 86.2%, 79.2% for H. pylori post-eradication images respectively.
While immunological tests can support the diagnosis of celiac disease, definitive diagnosis requires histological assessment of duodenal biopsies[72]. As such being able to identify changes in the duodenal mucosa consistent with celiac disease is important. However, these changes can be subtle and difficult to appreciate. Few studies have been published using a CADe system to detect or diagnose celiac disease.
In 2016, Gadermayr et al[73] created a system that combined expert knowledge acquisition with feature extraction to classify duodenal images obtained from 290 children as Marsh-0 (normal mucosa) or Marsh-3 (villous atrophy). Expert knowledge acquisition was achieved by having one of three study endoscopists assign a Marsh grade of 0 or 3 to an image. Feature extraction was accomplished using one of three methods: (1) multi-resolution local binary patterns; (2) multi-fractal spectrum; and (3) improved Fisher vectors. From expert knowledge acquisition and feature extraction, their classification algorithm identified images as Marsh-0 or Marsh-3. With optimal settings, the classification algorithm achieved an accuracy of 95.6%-99.6%[73]. In 2016, Wimmer et al[74] used CNN to detect celiac disease in a set of 1661 images (986 images of normal mucosa, 675 images of celiac disease) with varying convolutional blocks. Their CNN achieved the best overall classification rate (90.3%) with 4 convolutional blocks[74]. Taking their CNN a step further, they combined the CNN with 4 convolutional blocks with SVM which increased overall classification rate by 6.7%[74]. While interesting, Gadermayr et al’s method requires human intervention and the paper’s methodology is quite complicated[73], largely in part to the extensive number of systems tested. Wimmer et al[74] provided a simpler method that attained a good overall classification rate.
Few studies have assessed the utility of AI in the detection of celiac disease using WCE. In 2017, Zhou et al[75] trained GoogLeNet, a DCNN, to identify celiac disease using clips obtained during WCE. Their DCCN achieved a sensitivity and specificity of 100% for identifying patients with celiac disease from 10 WCE videos (5 from patients with celiac disease, 5 from healthy controls)[75]. Similarly, Wang et al[76] used DL to diagnose celiac disease from WCE videos, however their CNN utilized a block-wise channel squeeze and excitations attenuation module, a newer architectural unit thought to better mimic human visual perception[76]. Their system attained an accuracy of 95.9%, sensitivity of 97.2% and specificity of 95.6% for diagnosing celiac disease.
WCE is often used in patients with inflammatory bowel disease (IBD) to detect small bowel ulcers and erosions. While computed tomography enterography and MRI have been used to detect areas of disease activity and inflammation along the gastrointestinal tract in patients with IBD, these imaging modalities can miss early or small lesions. While WCE can directly visualize lesions, endoscopists reviewing the video may miss lesions or mistakenly identify imaging artifacts as lesions. AI systems could help reduce these errors. Several studies have been published using AI in WCE to detect intestinal changes consistent with Crohn’s disease[77-83].
To discriminate ulcers from normal mucosa in Crohn’s disease, Charisis et al[78] proposed combining bidimensional ensemble empirical mode decomposition and differential lacunarity to pre-process images followed by classification using several ML algorithms and a multilayer neural network. Using a dataset consisting of 87 ulcer and 87 normal mucosa images, their CADe achieved accuracy 89.0%-95.4%, sensitivity 88.2%-98.8%, and specificity 84.2%-96.6%[78]. Subsequently, Charisis and Hadjileontiadis published a paper in 2016 combining hybrid adaptive filtering and differential lacunarity (HAF-DLac) to process images followed by SVM to detect Crohn’s disease related lesions in WCE[79]. In a set of 800 WCE images, the HAF-DLac system achieved a sensitivity, specificity and accuracy of 95.2%, 92.4% and 93.8% respectively for detecting lesions[79]. Using a similar approach to Charisis et al[78], Kumar et al[80] used MPEG-7 edge, color and texture features to pre-process images followed by image classification using SVM to detect and classify lesions in patients with Crohn’s disease. Their system, tested against 533 images (212 normal mucosa, 321 images with lesions), obtained an accuracy of 93.0%-93.8% for detecting lesions and an accuracy of 78.5% for classifying them based on severity.
With respect to deep learning, few groups have used deep learning algorithms in WCE to identify Crohn’s disease related lesions. Recently, Ferreira et al[82] used a DCNN to identify erosions and ulcers in patients with Crohn’s disease. Their DCNN achieved a sensitivity of 98.0%, specificity of 99.0%, accuracy of 98.8% and AUROC of 1.00. Interestingly, Klang et al[83] developed a DCNN to detect intestinal strictures. Overall, their DCNN achieved an accuracy of 93.5% ± 6.7% and AUC of 0.989 for detecting strictures.
Three studies have used artificial intelligence to detect hookworms using WCE. The first to publish on this topic was Wu et al[84] in 2016. Using SVM, they were able to create a system that achieved a specificity of 99.0% and accuracy of 98.4% for detecting hookworms in WCE[84]. However, the system’s sensitivity was 11.1%. He et al[85] created a DCNN using a novel deep hookworm detection framework that modeled the tubular appearance of hookworms. Their DCNN had an accuracy of 88.5% for identifying hookworm[85]. Gan et al[86] performed a similar study, finding an AUC of 0.97 (95%CI 0.967-0.978), sensitivity of 92.2%, specificity of 91.1% and accuracy of 91.2% The concordant findings of these three studies suggest a possible utility of using AI to diagnose hookworm infections.
One of the most common reasons to perform WCE is to evaluate for gastrointestinal bleeding after prior endoscopic attempts have failed to localize a source. Since the implementation of WCE in clinical practice, many methods, notably AI, have been employed to improve the detection of gastrointestinal sources of bleeding.
Several studies have looked at using supervised learning to identify bleeding in WCE. In 2014, Sainju et al[87] used an ML algorithm to interpret color quantization images and determine if bleeding was present. One of their models achieved a sensitivity, specificity and accuracy of 96%, 90% and 93%, respectively[87]. Using SVM, Usman et al[88] achieved similar results - sensitivity, specificity and accuracy of 94%, 91% and 92% respectively.
More recently, several groups have created DCNNs to identify bleeding and sources of bleeding in WCE. In 2021, Ghosh et al[89] used a system comprised of two CNN systems (CNN-1, CNN-2) to classify WCE images as bleeding or non-bleeding and subsequently to identify sources of bleeding within the bleeding images. For classifying images as bleeding or non-bleeding, CNN-1 had a sensitivity, specificity, accuracy and AUC of 97.5%, 99.9%, 99.4% and 0.99[89]. For identifying sources of bleeding within the bleeding images, CNN-2 had an accuracy of 94.4% and intersection over union (IoU) of 90.7%[89].
In 2020, Tsuboi et al[90] published the first study to use DCNN to detect small bowel angioectasias from WCE images. In their test set which included 488 images of small bowel angioectasias and 10000 images of normal small bowel mucosa, their DCNN achieved an AUC of 0.99 with sensitivity and specificity of 98.8% and 98.4%[90]. Similarly, in 2021 Ribeiro at al[91] developed a DCNN to identify vascular lesions, categorizing them by bleeding risk according to Saurin’s classification: P0 – no hemorrhagic potential, P1 – uncertain/intermediate hemorrhagic potential and red spots, and P2 – high hemorrhagic potential (angioectasias, varices). In their validation set, the DCNN had a sensitivity, specificity, accuracy and AUROC of 91.7%, 95.3%, 94.1% and 0.97 respectively for identifying P1 lesions[91]. Regarding P2 lesions, the network had a sensitivity, specificity, accuracy and AUROC of 94.1%, 95.1%, 94.8% and 0.98 respectively[91]. This group published a similar study in 2022 however now using their DCNN to detect and differentiate mucosal erosions and ulcers based on bleeding potential[92]. Saurin’s classification was again used to classify lesions, additionally labeling P1 lesions as mucosal erosions or small ulcers and P2 lesions as large ulcers (> 2 cm)[92]. The DCNN achieved an overall sensitivity of 90.8% ± 4.7%, specificity of 97.1% ± 1.7%, and accuracy of 93.4% ± 3.3% in their test set of 1226 images[92]. For the detection of mucosal erosions (P1), their DCNN achieved a sensitivity of 87.2%, specificity of 95.0% and accuracy of 93.3% with AUROC of 0.98 (95%CI 0.97-0.99)[92]. With respect to small ulcers (P1), their DCNN achieved a sensitivity of 86.4%, specificity of 96.9% and accuracy of 94.5% with AUROC of 0.99 (95%CI 0.97-1.00)[92]. Finally, with respect to large ulcers (P2), their DCNN achieved a sensitivity of 95.3%, specificity of 99.2% and AUROC of 1.00 (95%CI 0.98-1.00)[92]. A third study published by this group aimed to develop a DCNN to identify colonic lesions and luminal blood/hematic vestiges had similar findings. In their training set of 1801 images, the DCNN achieved an overall sensitivity, specificity and accuracy of 96.3%, 98.2%, and 97.6% respectively[93]. For detecting mucosal lesions, the DCNN achieved a sensitivity of 92.0%, specificity of 98.5% and AUROC of 0.99 (95%CI 0.98-1.00)[93]. For luminal blood/hematic vestiges, the DCNN achieved a sensitivity of 99.5%, specificity of 99.8% and AUROC of 1.00 (95%CI 0.99-1.00)[93].
Gastrointestinal tumors can be difficult to discern from normal mucosa and thus pose a higher degree of diagnostic difficulty compared to other lesions on traditional WCE[94]. As such, developing an AI system to aid with the detection of these easy to miss lesions could be beneficial.
Several groups have developed ML systems to aid with detection. Using SVM, Li et al[95] were able to develop a system capable of detecting small bowel tumors with sensitivity, specificity and accuracy of 88.6%, 96.2% and 92.4%. Similarly, Liu et al[96] and Faghih Dinevari et al[97] used SVM to identify tumors in WCE, however they used different image pre-processing algorithms. Liu et al[96] used discrete curvelet transform to pre-process images prior to being classified by SVM. Their ML system achieved a sensitivity of 97.8% ± 0.5, specificity of 96.7% ± 0.4 and accuracy of 97.3% ± 0.5 for identifying small bowel tumors[96]. Faghih Dinevari et al[97] relied on discrete wavelet transform and singular value decomposition for image pre-processing prior to classification by SVM. Their system achieved a sensitivity of 94.0%, specificity of 93.0% and accuracy of 93.5% for identifying small bowel tumors[97]. Sundaram and Santhiyakumari built upon these methodologies, using a region of interest-based color histogram to enhance WCE images prior to being classified by two SVM algorithms: SVM1 and SVM2[98]. SVM1 classified the WCE image as normal or abnormal. If SVM1 classified the image as abnormal, it was further classified by SVM2 as benign, malignant or normal[98]. The system attained an overall sensitivity of 96.0%, specificity of 95.4% and accuracy of 95.7% for small bowel tumor detection and classification[98].
With respect to DL methods, Blanes-Vidal et al[99] created a DCNN to autonomously detect and localize colorectal polyps. Their study included 255 patients who underwent WCE and standard colonoscopy for positive fecal immunochemical tests. Of the 255 patients, 131 had at least 1 polyp. The DCNN obtained a sensitivity of 97.1%, specificity of 93.3% and accuracy of 96.4% for detecting polyps in WCE[99]. Saraiva et al[100] and Mascarenhas et al[101] similarly used DCNNs to detect colonic polyps in WCE and obtained similar results to Blanes-Vidal et al[99-101]. Using an ANN, Constantinescu et al[102] created a DL system able to detect small bowel polyps with sensitivity of 93.6% and specificity of 91.4%. For gastric polyps and tumors, Xia et al[103] created a novel CNN – a region-based convolutional neural network (RCNN) – to evaluate magnetically controlled capsule endoscopy (MCE) images. Tested on 201365 MCE images obtained from 100 patients, the RCNN detected gastric polyps with sensitivity of 96.5%, specificity of 94.8%, accuracy of 94.9% and AUC of 0.898 (95%CI 0.84-0.96)[103]. For submucosal tumors, the RCNN achieved a sensitivity of 87.2%, specificity of 95.3%, accuracy of 95.2% and AUC of 0.88 (95%CI 0.81-0.96)[103]. Taking a different approach, Yuan and Meng used a novel deep learning method – stacked sparse autoencoder image manifold constraint – to identify intestinal polyps on WCE, finding an accuracy of 98.00% for poly detection[104]. However, sensitivity, specificity and AUC analyses were not reported.
Inadequate bowel preparation, present in 15% to 35% of colonoscopies, is associated with lower rates of cecal intubation, lower adenoma detection rate (ADR), and higher rates of procedure-related adverse events[105,106]. For patients with inadequate bowel preparation, the United States Multi-Society Task Force of Colorectal Cancer (MSTF) which represents the American College of Gastroenterology, the American Gastroenterological Association and the American Society for Gastrointestinal Endoscopy (ASGE), and the European Society of Gastrointestinal Endoscopy recommend repeating a colonoscopy within 1 year[105,107-109]. In addition, the MSTF and ASGE recommend that endoscopists document bowel preparation quality at time of colonoscopy[108,109].
Despite these recommendations and variety of bowel preparation rating scales available, documentation of bowel preparation quality remains variable with studies reporting appropriate documentation in 20% to 88% of colonoscopies[110-112]. Few studies have been published regarding the use of DCNN to assist in the objective assessment of bowel preparation. The first group to do so, Zhou et al[113] in 2019, found that their DCNN (ENDOANGEL) was more accurate (93.3%) at grading the bowel preparation quality of still images than novice (< 1 year of experience performing colonoscopies; 75.91%), senior (1-3 years of experience performing colonoscopies; 74.36%) and expert (> 3 years of experience performing colonoscopies; 55.11%) endoscopists. When tested on colonoscopy videos, ENDOANGEL remained accurate at grading bowel preparation quality (89.04%)[113].
Building upon their experience with ENDOANGEL, Zhou et al[114] created a new system using two DCNNs: DCNN1 filtered unqualified frames while DCNN2 classified images by Boston Bowel Preparation Scale (BBPS) scores. The BBPS is a validated rating scale for assessing bowel preparation quality[115]. Colonic segments are assigned scores on a scale from 0 to 3. Colonic segments unable to be evaluated due to the presence of solid, unremovable stool are assigned a score of 0 whereas colonic segments that are able to be easily evaluated and contain minimal to no stool are assigned a score of 3[115]. Zhou et al’s DCNN2 classified images into two categories: well-prepared (BBPS score 2-3) and poorly prepared (BPPS score 0-1)[114]. There was no difference between the dual DCNN system and study endoscopists when calculating the unqualified image portion (28.35% vs 29.58%, P = 0.285) and e-BBPS scores (7.81% vs 8.74%, P = 0.088). In addition, a strong inverse relationship between e-BBPS and ADR (ρ = -0.976, P = 0.022) was found.
Two other groups developed similar dual DCNN systems as Zhou et al[114] to calculate BBPS and obtained concordant findings[116,117]. Lee et al[116] tested their system on colonoscopy videos and found the system had an accuracy of 85.3% and AUC of 0.918 for detecting adequate bowel preparation. Using still images, Low et al’s system was able to accurately determine bowel preparation adequacy (98%) and subclassify by BBPS (91%)[117].
Using a different approach, Wang et al[118] used U-Net to create a DCNN to perform automatic segmentation of fecal matter from still images. Compared to images segmented by endoscopists, U-Net achieved an accuracy of 94.7%.
Colonoscopy is essential for the assessment of IBD as it allows for real-time evaluation of colonic inflammation[119,120]. Despite there being endoscopic scoring systems available to quantify disease activity, assessment is operator-dependent resulting in high interobserver variability[119-121]. Recent efforts have focused on using artificial intelligence to objectively grade colonic inflammation[121,122].
Several studies have investigated using DCNNs to classify images obtained from patients with ulcerative colitis (UC) by endoscopic inflammation scoring systems. The most commonly used endoscopic scoring system in these studies is the Mayo Endoscopic Score (MES). Physicians assign scores on a scale from 0 to 3 based on the absence or presence of erythema, friability, erosions, ulceration and bleeding[123]. A score of 0 indicates normal or inactive mucosa whereas a score of 3 indicates severe disease activity[123]. In 2018, Ozawa et al[121] published the first study to use a DCNN to classify still images obtained from patients with UC into MES 0 vs MES 1-3 and MES 0-1 vs MES 2-3. Their DCNN had an AUROC of 0.86 (95%CI 0.84-0.87) and AUROC 0.98 (95%CI 0.97-0.98) when differentiating MES 0 vs MES 1-3 and MES 0-1 vs MES 2-3 respectively[121]. Stidham et al[122] performed a similar study and found an AUROC of 0.966 (95%CI 0.967-0.972) for differentiating still images into MES 0-1 vs MES 2-3. Using a combined deep learning and machine learning system, Huang et al[124] were able to achieve an AUC of 0.938 with accuracy of 94.5% for identifying MES 0-1 vs MES 2-3 from still images. While the binary classification used in the aforementioned studies can differentiate remission/mucosal healing (MES 0-1) and active inflammation (MES 2-3), knowing exact MESs also has clinical significance[125,126]. Bhambhvani and Zamora created a DCNN to assign individual MESs to still images. The model achieved an AUC of 0.89, 0.86 and 0.96 for classifying images into MES 1, MES 2 and MES 3 respectively and achieved an average specificity of 85.7%, average sensitivity of 72.4% and overall accuracy of 77.2%[127].
In order to simulate how MES is performed in practice, several groups developed systems using DL to predict MES from colonoscopy videos. Yao et al’s DCNN had good agreement with MES scoring performed by gastroenterologists in their internal video test set (k = 0. 84; 95%CI 0.75-0.92), however their DCCN did not perform as well in the external video test set (k = 0.59; 95%CI 0.46-0.71)[128]. Gottlieb et al[129] reported similar findings to Yao et al[128], finding that their DCNN had good agreement with MES scoring performed by gastroenterologists (quadratic weighted kappa of 0.844; 95%CI 0.787–0.901). Gutierrez Becker et al[130] created a DL system designed to perform multiple binary tasks: discriminating MES < 1 vs MES ≥ 1, MES < 2 vs MES ≥ 2, and MES < 3 vs MES ≥ 3. For these tasks, their DL system attained an AUROC of 0.84, 0.85, and 0.85 respectively.
A group from Japan published several studies using AI on endoscopic images to predict histologic activity in patients with UC[131-134]. Their first study in 2016 used machine learning to predict persistent histologic inflammation[131]. Their system attained a sensitivity of 74% (95%CI 65%-81%), specificity of 97% (95%CI 95%-99%) and accuracy of 91% (95%CI 83%-95%) for predicting persistent histologic inflammation in still images[131]. Their following studies used a deep neural network labeled DNUC (deep neural network for evaluation of UC) to identify endoscopic remission and histologic remission[132,134]. In still images, DNUC had a sensitivity of 93.3% (95%CI 92.2%–94.3%), specificity of 87.8% (95%CI 87.0%–88.4%) and diagnostic accuracy of 90.1% (95%CI 89.2%–90.9%) for determining endoscopic remission[132]. With respect to histologic remission, DNUC had a sensitivity of 92.4% (95%CI 91.5%–93.2%), specificity of 93.5% (95%CI 92.6%–94.3%) and diagnostic accuracy of 90.1% (92.9%; 95%CI 92.1%–93.7%)[132]. In colonoscopy videos, DNUC showed a sensitivity of 81.5% (95%CI 78.5%–83.9%) and specificity of 94.7% (95%CI 92.5%–96.4%) for endoscopic remission[134]. For histologic remission, DNUC had a sensitivity of 97.9% (95%CI 97.0%–98.5%) and specificity of 94.6% (95%CI 91.1%–96.9%) in colonoscopy videos[134].
To date, only one study has been published using an AI system to distinguish normal from inflamed colonic mucosa in Crohn’s disease[135]. The group paired a DCNN with a long short-term memory (LSTM), a type of neural network that uses previous findings to interpret its current input, and confocal laser endomicroscopy. Their DCNN-LSTM system attained an accuracy of 95.3% and AUC of 0.98 for differentiating normal from inflamed mucosa[135].
Colorectal cancer is the third most common malignancy and second leading cause of cancer-related mortality in the world[136]. While colonoscopy is the gold standard for detection and treatment of premalignant and malignant lesions, a substantial number of adenomas are missed[137,138]. As such, efforts have focused on using AI to improve ADR and decrease adenoma miss rate (AMR).
At present, numerous pilot, validation and prospective studies[139-161], randomized controlled studies[162-174], and systematic reviews and meta-analyses[175-183] have been published regarding the use of AI for the detection of colonic polyps. Furthermore, there are commercially available AI systems for both polyp detection and interpretation. With respect to the systematic reviews and meta-analyses published on this topic, AI-assisted colonoscopy has consistently been shown to have higher ADR, polyp detection rate (PDR) and adenoma per colonoscopy (APC) compared to standard colonoscopy[175-183]. Recently, several large, randomized controlled trials have been published supporting these findings. Shaukat et al[162] published their findings from their multicenter, randomized controlled trial comparing CADe colonoscopy to standard colonoscopy. Their study included 1359 patients: 677 randomized to standard colonoscopy, 682 to CADe colonoscopy. They found an increase in ADR (47.8% vs 43.9%; P = 0.065) and APC (1.05 vs 0.83; P = 0.002) in the CADe colonoscopy group. However, they also found a decrease in the overall sessile serrated lesions per colonoscopy rate (0.20 vs 0.28; P = 0.042) and sessile serrated lesion detection rate (12.6% vs 16.0%; P = 0.092) in the CADe colonoscopy group[162]. Brown et al[163] in their CADeT-CS Trial which was a multicenter, single-blind randomized tandem colonoscopy study comparing CADe colonoscopy to high-definition white light colonoscopy found similar increases in ADR (50.44% vs 43.64%; P = 0.3091) and APC (1.19 vs 0.90; P = 0.0323) in their patients who underwent CADe colonoscopy first[163]. Additionally, polyp miss rate (PMR) (20.70% vs 33.71%; P = 0.0007), AMR (20.12% vs 31.25%; P = 0.0247), and sessile serrated lesion miss rate (7.14% vs 42.11%; P = 0.0482) were lower in the CADe colonoscopy first group. In a similarly designed study to Brown et al[163], Kamba et al’s multicenter, randomized tandem colonoscopy study comparing CADe colonoscopy to standard colonoscopy found lower AMR (13.8% vs 26.7%; P < 0.0001), PMR (14.2% vs 40.6%; P < 0.0001), and sessile serrated lesion miss rate (13.0% vs 38.5%’ P = 0.03) and higher ADR (64.5% vs 53.6%; P = 0.036) and PDR (69.8% vs 60.9%; P = 0.084) in patients who underwent CADe colonoscopy first[164]. Similar to Shaukat et al[162], the sessile serrated lesion detection rate was lower in the CADe colonoscopy first group compared to standard colonoscopy first (7.6% vs 8.1%; P = 0.866)[164]. Similar increases in ADR, APC and PDR were appreciated in randomize controlled trials by Xu et al[172], Liu et al[173], Repici et al[170], Gong et al[166], Wang et al[167], and Su et al[169] as well[166-172].
The majority of AI-assisted colonoscopy studies focus on adenoma detection. While these studies report sessile serrated lesion rates, it is often a secondary outcome despite sessile serrated lesions being the precursors of 15%-30% of all colorectal cancers[184]. Few studies have created AI systems optimized for dedicating sessile serrated lesions. Recently, Yoon et al[184] used a generative adversarial network (GAN) to generate endoscopic images of sessile serrated lesions which were used to train their DCNN with the hope of improving sessile serrated lesion detection. In the validation set which was comprised of 1141 images of polyps and 1000 normal images, their best performing GAN-DCNN model, GAN-aug2, achieved a sensitivity of 95.44% (95%CI 93.71%-97.17%), specificity of 90.10% (95%CI 88.38%-91.77%), accuracy of 92.95% (95%CI 91.86%-94.04%) and AUROC of 0.96 (95%CI 0.9547-0.9709)[184]. In a type-separated polyp validation dataset, the GAN-aug2 achieved a sensitivity of 95.24%, 19.1% higher than the DCNN without augmentation[184]. Given the small number of sessile serrate lesions present in the initial set, Yoon et al[184] collected an additional 130 images depicting 133 sessile serrated lesions to create an additional validation set titled SSL temporal validation dataset[184]. The GAN-aug2 continued to outperform the DCNN without augmentation (sensitivity 93.98% vs 84.21%). Nemoto et al[185] created a DCNN to differentiate (1) tubular adenomas from serrated lesions; and (2) serrated lesions from hyperplastic polyps. In their 215-image training set, the DCNN was able to differentiate tubular adenomas from sessile serrated lesions with sensitivity of 72% (95%CI 62%-81%), specificity 89% (95%CI 82%-94%), accuracy 82% (95%CI 77%-87%) and AUC 0.86 (95%CI 0.80-0.91). For differentiating sessile serrated lesions from hyperplastic polyps, the DCNN achieved a sensitivity of 17% (95%CI 7%-32%), specificity 85% (95%CI 76%-92%), accuracy 63% (95%CI 54%-72%) and AUC 0.55 (95%CI 0.44-0.66)[185]. An overview of studies investigating the detection accuracy of CADe is provided in Table 3. An overview of studies investigating ADR and PDR using CADe is provided in Table 4.
| Ref. | Country | Study design | Lesions | Training dataset | Test dataset | Sensitivity (%) | Specificity (%) | Accuracy (%) | AUROC |
| Komeda et al[139], 2017 | Japan | Retrospective | Adenomas | 1200 images | 10 images | 80 | 60 | 70 | - |
| Misawa et al[140], 2018 | Japan | Retrospective | Polyps | 411 video clips | 135 video clips | 90 | 63.3 | 76.5 | 0.87 |
| Wang et al[149], 2018 | China, United States | Retrospective | Polyps | 4495 images | Dataset A: 27113 images; Dataset C: 138 video clips; Dataset D: 54 full-length videos | Dataset A: 94.38; Dataset C: 91.64 | Dataset A: 95.92; Dataset D: 95.4 | - | Dataset A: 0.984 |
| Horiuchi et al[154], 2019 | Japan | Prospective | Diminutive polyps | - | a | 80 | 95.3 | 91.5 | - |
| Hassan et al[141], 2020 | Italy, United States | Retrospective | Polyps | - | 338 video clips | 99.7 | - | - | - |
| Guo et al[142], 2021 | Japan | Retrospective | Polyps | 1991 images | 100 video clips; 15 full videos | 87b | 98.3b | - | - |
| Neumann et al[143], 2021 | Germany | Retrospective1 | Polyps | > 500 videos | 240 polyps within full-length videos | 100 | 0 | - | - |
| Li et al[144], 2021 | Singapore | Retrospective | Polyps | 6038 images | 2571 images | 74.1 | 85.1 | - | - |
| Livovsky et al[151], 2021 | Israel | Ambispective | Polyps | 3611 h of videos | 1393 h of videos | 97.1 | 0 | - | - |
| Pfeifer et al[158], 2021 | Germany, Italy, Netherlands | Retrospective | Polyps | 10467 images | 45 videos | 90 | 80 | - | 0.92 |
| Ahmad et al[145], 20222 | England | Prospective | Polyps | Dataset A: 58849 frames; Dataset B: 10993 videos and still images | Dataset C: 110985 frames; Dataset D: 8950 frames; Dataset E: 542484 frames | Dataset C: 100, 84.1; Dataset D&E: 98.9, 85.2 | Dataset C: 79.6; Dataset D&E: 79.3% | ||
| Hori et al[146], 2022 | Japan | Prospective | Polyps | 1456 images | 600 images | 97 | 97.7 | 97.3 | - |
| Pacal et al[152], 2022 | Turkey | Retrospective | Polyps | Used images from 3 publicly available datasets (SUN, PICCOLO, Etis-Larib) to create training and test datasets | 91.04 | - | - | - | |
| Yoon et al[184], 2022 | South Korea | Retrospective | SSL | 4397 images | Validation Set 2106; SSL Temporal Validation set 133 | 95.44; 93.89 | 90.1 | 92.95 | 0.96 |
| Nemoto et al[185], 2022 | Japan | Retrospective | TA, SSL | 1849 images | 400 images | 72 | 89 | 82 | 0.86 |
| Lux et al[148], 2022 | Germany | Retrospective | Polyps | 506338 images | 41 full-length videos | - | - | 95.3 | - |
| Ref. | Country | Study design | Patients (n) | PDR (%) | ADR (%) | |||||
| CADe | SC | CADe | SC | P value | CADe | SC | P value | |||
| Wang et al[168], 2019 | China, United States | Randomized | 522 | 536 | 45.02 | 29.1 | < 0.001 | 29.12 | 20.34 | < 0.001 |
| Becq et al[155], 2020 | United States, Turkey, Costa Rica | Prospective | 50b | 82 | 62 | Not reported | - | - | - | |
| Gong et al[166], 2020 | China | Randomized | 355 | 349 | 47 | 34 | 0.0016 | 16 | 8 | 0.001 |
| Liu et al[171], 2020 | China, United States | Randomized | 393 | 397 | 47.07 | 33.25 | < 0.001 | 29.01 | 20.91 | 0.009 |
| Liu et al[173], 2020 | China | Prospective | 508 | 518 | 43.65 | 27.81 | < 0.001 | 39.1 | 23.89 | < 0.001 |
| Repici et al[170], 2020 | Italy, Kuwait, United States, Germany | Randomized | 341 | 344 | - | - | - | 54.8 | 40.4 | < 0.001 |
| Su et al[169], 2020 | China | Randomized | 308 | 315 | 38.3 | 25.4 | 0.001 | 28.9 | 16.5 | < 0.001 |
| Wang et al[156], 2020 | China, United States | Prospective, Tandem1 | 184 | 185 | 65.59 | 55.14 | 0.099 | 42.39 | 35.68 | 0.186 |
| Wang et al[167], 2020 | China, United States | Randomized | 484 | 478 | 52 | 37 | < 0.0001 | 34 | 28 | 0.03 |
| Kamba et al[164], 2021 | Japan | Randomized, Tandem2 | 172 | 174 | 69.8 | 60.9 | 0.084 | 64.5 | 53.6 | 0.036 |
| Luo et al[174], 2021 | China | Randomized, Tandem1 | 72 | 78 | 38.7 | 34 | < 0.001 | - | - | - |
| Pfeifer et al[158], 2021 | Germany, Italy, Netherlands | Prospective, Tandem1 | 42b | 50 | 38 | 0.023 | 36 | 26 | 0.044 | |
| Shaukat et al[157], 2021 | United States, England | Prospective | 83 | 283 | - | - | - | 54.2 | 40.6 | 0.028 |
| Shen et al[150], 2021 | China | Ambispective | 64 | 64 | 78.1 | 56.3 | 0.008 | 53.1 | 29.7 | 0.007 |
| Xu et al[172], 2021 | China | Randomized | 1177 | 1175 | 38.8 | 36.2 | 0.183 | - | - | - |
| Glissen Brown et al[163], 2022 | China, United States | Randomized, Tandem2 | 113 | 110 | 70.8 | 65.45 | 0.3923 | 50.44 | 43.64 | 0.3091 |
| Ishiyama et al[159], 2022 | Japan, Norway | Prospective | 918 | 918 | 59 | 52.1 | 0.003 | 26.4 | 19.9 | 0.001 |
| Lux et al[148], 2022 | Germany | Retrospective | 41 | - | - | - | - | - | 41.5 | - |
| Quan et al[153], 2022 | United States | Prospective | 300 | 300 | - | - | - | 43.7a; 66.7 | 37.8a; 59.72 | 0.37a; 0.35 |
| Repici et al[165], 2022 | Italy, Switzerland, United States, Germany | Randomized | 330 | 330 | - | - | - | 53.3 | 44.5 | 0.017 |
| Shaukat et al[162], 2022 | United States | Randomized | 682 | 677 | 64.4 | 61.2 | 0.242 | 47.8 | 43.9 | 0.065 |
| Zippelius et al[160], 2022 | Germany, United States | Prospective | 150b | - | - | - | 50.7 | 52 | 0.5 | |
Artificial intelligence is in its early stages for medicine, especially in gastroenterology and endoscopy. AI will help is in the areas of “augmentation” and “automation”. Augmentation like what is happening with polyp detection and interpretation. Automation by eliminating electronic paperwork, such as the use of natural language processing for procedure documentation. Artificial intelligence systems have repeatedly been shown to be effective at identifying gastrointestinal lesions with high sensitivity, specificity and accuracy. While lesion detection is important, this is only the beginning of AI’s utility in esophagogastroduodenoscopy, WCE and colonoscopy.
After refining their AI systems for lesion detection, several groups discussed in this narrative review were able to add additional functions to their AI systems. In BE, ESCC and gastric cancer, several AI systems were capable of predicting tumor invasion depth. Within IBD, AI systems were able to generate endoscopic disease severity scores. One group was able to train their CADe to recommend neoplasia biopsy sites in BE[14]. Additional efforts should be dedicated to developing these functions, testing them in real-time and having the AI system provide management recommendations when clinically appropriate.
Additional areas in need of future research are using AI systems to make histologic predictions, to assist with positioning of the endoscopic ultrasound (EUS) transducer and interpretation of EUS images, to detect biliary diseases and make therapeutic recommendations in endoscopic retrograde cholangiopancreatography (ERCP), and, in combination with endoscopic mechanical attachments, to improve colorectal cancer screening and surveillance. While endoscopists may perform optical biopsies of gastrointestinal lesions to predict histology and make real-time management decisions, these predictions are highly operator-dependent and often require expensive equipment that is not readily available. Thus, developing an AI system capable of performing objective optical biopsies, especially in WLE, would preserve the quality of histologic predictions, be cost effective, and avoid the risks associated with endoscopic biopsy and resection.
Similarly, EUS is highly operator-dependent, requiring endoscopists to place the transducer in specific positions to obtain adequate views of the hepatopancreatobiliary system. Research should focus on using AI systems to assist with appropriate transducer positioning and perform real-time EUS image analysis[186-194].
Presently, several clinical studies are actively recruiting patients to evaluate the utility of AI systems in ERCP. Of particular interest is the diagnosis and management of biliary diseases. Some groups are planning to use AI to classify bile duct lesions and provide biopsy site recommendations[195]. One group is planning to use an AI system in patients requiring biliary stents to assist with biliary stent choice and stent placement[196]. It will be interesting to see how AI performs in these tasks as successes could pave the way for future studies investigating the utility of AI systems to make real-time management recommendations.
While this narrative review focused on the use of AI in colonoscopy, of growing interest is the use of endoscopic mechanical attachments in colonoscopy to assist with polyp detection in colorectal cancer screening and surveillance. Independently, AI systems and endoscopic mechanical attachments are known to increase ADR and PDR. Few studies have investigated how combining AI with endoscopic mechanical attachments impacts ADR and PDR. Future research should examine the impact that combining these modalities has on ADR and PDR.
While substantial advances have been made in AI, it is important to note that AI is not without limitations. In many of the studies discussed in this narrative review, the authors trained their AI systems using internally obtained images labeled by a single endoscopist. Thus, the AI is subject to the same operator biases and human error as the labeling endoscopist[1,197]. In addition, by using internally obtained data, several of these training sets may have inherent institutional or geographic biases resulting in AI systems that are biased and nongeneralizable[197]. As AI continues to progress, large datasets comprised of high-quality images should be created and used for training AI systems to reduce these biases[1].
With the implementation of AI in clinical practice, medical error accountability must also be addressed. While many of the AI systems discussed in this narrative review boast high detection accuracies, none are perfect. It is undeniable that errors in detection and diagnosis will arise when using these technologies. Regulatory bodies are needed to continually supervise these AI systems and oversee problems as they arise[198].
In this narrative review, we provide an objective overview of the AI-related research being performed within esophagogastroduodenoscopy, WCE and colonoscopy. We attempted to be comprehensive by using several electronic databases including Embase, Ovid Medicine, and PubMed. However, it is possible that some publications pertinent to our narrative review were missed.
Undoubtedly, AI within esophagogastroduodenoscopy, WCE and colonoscopy is rapidly evolving, moving from retrospectively tested supervised learning algorithms to large, multicenter clinical trials using completely autonomous systems within the span of 10 years. The systems developed by these researchers show promise for detecting lesions, diagnosing conditions, and monitoring diseases. In fact, two of the computer aided detection systems discussed in this narrative review designed to aid with colorectal polyp detection were approved by the United States Food and Drug Administration in 2021[171,199]. Thus, the question is no longer if but when will AI become integrated with clinical practice. Medical providers at all levels of training should prepare to incorporate artificial intelligence systems into routine practice.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Liu XQ, China; Qi XS, China S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
| 1. | Kröner PT, Engels MM, Glicksberg BS, Johnson KW, Mzaik O, van Hooft JE, Wallace MB, El-Serag HB, Krittanawong C. Artificial intelligence in gastroenterology: A state-of-the-art review. World J Gastroenterol. 2021;27:6794-6824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 28] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (7)] |
| 2. | Kaul V, Enslin S, Gross SA. History of artificial intelligence in medicine. Gastrointest Endosc. 2020;92:807-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 314] [Article Influence: 62.8] [Reference Citation Analysis (1)] |
| 3. | Deo RC. Machine Learning in Medicine. Circulation. 2015;132:1920-1930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1155] [Cited by in RCA: 1956] [Article Influence: 217.3] [Reference Citation Analysis (6)] |
| 4. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology. 2020;158:76-94.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 323] [Article Influence: 64.6] [Reference Citation Analysis (1)] |
| 5. | Christou CD, Tsoulfas G. Challenges and opportunities in the application of artificial intelligence in gastroenterology and hepatology. World J Gastroenterol. 2021;27:6191-6223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (8)] |
| 6. | Pannala R, Krishnan K, Melson J, Parsi MA, Schulman AR, Sullivan S, Trikudanathan G, Trindade AJ, Watson RR, Maple JT, Lichtenstein DR. Artificial intelligence in gastrointestinal endoscopy. VideoGIE. 2020;5:598-613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 7. | Hussein M, González-Bueno Puyal J, Mountney P, Lovat LB, Haidry R. Role of artificial intelligence in the diagnosis of oesophageal neoplasia: 2020 an endoscopic odyssey. World J Gastroenterol. 2020;26:5784-5796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Sharma P. Barrett Esophagus: A Review. JAMA. 2022;328:663-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 54] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 10. | van der Sommen F, Zinger S, Curvers WL, Bisschops R, Pech O, Weusten BL, Bergman JJ, de With PH, Schoon EJ. Computer-aided detection of early neoplastic lesions in Barrett's esophagus. Endoscopy. 2016;48:617-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (2)] |
| 11. | Swager AF, van der Sommen F, Klomp SR, Zinger S, Meijer SL, Schoon EJ, Bergman JJGHM, de With PH, Curvers WL. Computer-aided detection of early Barrett's neoplasia using volumetric laser endomicroscopy. Gastrointest Endosc. 2017;86:839-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 12. | Struyvenberg MR, van der Sommen F, Swager AF, de Groof AJ, Rikos A, Schoon EJ, Bergman JJ, de With PHN, Curvers WL. Improved Barrett's neoplasia detection using computer-assisted multiframe analysis of volumetric laser endomicroscopy. Dis Esophagus. 2020;33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | de Groof J, van der Sommen F, van der Putten J, Struyvenberg MR, Zinger S, Curvers WL, Pech O, Meining A, Neuhaus H, Bisschops R, Schoon EJ, de With PH, Bergman JJ. The Argos project: The development of a computer-aided detection system to improve detection of Barrett's neoplasia on white light endoscopy. United European Gastroenterol J. 2019;7:538-547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 14. | de Groof AJ, Struyvenberg MR, van der Putten J, van der Sommen F, Fockens KN, Curvers WL, Zinger S, Pouw RE, Coron E, Baldaque-Silva F, Pech O, Weusten B, Meining A, Neuhaus H, Bisschops R, Dent J, Schoon EJ, de With PH, Bergman JJ. Deep-Learning System Detects Neoplasia in Patients With Barrett's Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology. 2020;158:915-929.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 222] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 15. | de Groof AJ, Struyvenberg MR, Fockens KN, van der Putten J, van der Sommen F, Boers TG, Zinger S, Bisschops R, de With PH, Pouw RE, Curvers WL, Schoon EJ, Bergman JJGHM. Deep learning algorithm detection of Barrett's neoplasia with high accuracy during live endoscopic procedures: a pilot study (with video). Gastrointest Endosc. 2020;91:1242-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 16. | Struyvenberg MR, de Groof AJ, van der Putten J, van der Sommen F, Baldaque-Silva F, Omae M, Pouw R, Bisschops R, Vieth M, Schoon EJ, Curvers WL, de With PH, Bergman JJ. A computer-assisted algorithm for narrow-band imaging-based tissue characterization in Barrett's esophagus. Gastrointest Endosc. 2021;93:89-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 17. | Jisu Hong, Bo-Yong Park, Hyunjin Park. Convolutional neural network classifier for distinguishing Barrett's esophagus and neoplasia endomicroscopy images. Annu Int Conf IEEE Eng Med Biol Soc. 2017;2017:2892-2895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 18. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Prinz F, de Souza LA Jr, Papa J, Palm C, Messmann H. Real-time use of artificial intelligence in the evaluation of cancer in Barrett's oesophagus. Gut. 2020;69:615-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 124] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 19. | Hashimoto R, Requa J, Dao T, Ninh A, Tran E, Mai D, Lugo M, El-Hage Chehade N, Chang KJ, Karnes WE, Samarasena JB. Artificial intelligence using convolutional neural networks for real-time detection of early esophageal neoplasia in Barrett's esophagus (with video). Gastrointest Endosc. 2020;91:1264-1271.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 144] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 20. | Hussein M, González-Bueno Puyal J, Lines D, Sehgal V, Toth D, Ahmad OF, Kader R, Everson M, Lipman G, Fernandez-Sordo JO, Ragunath K, Esteban JM, Bisschops R, Banks M, Haefner M, Mountney P, Stoyanov D, Lovat LB, Haidry R. A new artificial intelligence system successfully detects and localises early neoplasia in Barrett's esophagus by using convolutional neural networks. United European Gastroenterol J. 2022;10:528-537. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 21. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Souza LA Jr, Papa JP, Palm C, Messmann H. Computer-aided diagnosis using deep learning in the evaluation of early oesophageal adenocarcinoma. Gut. 2019;68:1143-1145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 22. | Ali S, Bailey A, Ash S, Haghighat M; TGU Investigators, Leedham SJ, Lu X, East JE, Rittscher J, Braden B. A Pilot Study on Automatic Three-Dimensional Quantification of Barrett's Esophagus for Risk Stratification and Therapy Monitoring. Gastroenterology. 2021;161:865-878.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 23. | Ebigbo A, Mendel R, Rückert T, Schuster L, Probst A, Manzeneder J, Prinz F, Mende M, Steinbrück I, Faiss S, Rauber D, de Souza LA Jr, Papa JP, Deprez PH, Oyama T, Takahashi A, Seewald S, Sharma P, Byrne MF, Palm C, Messmann H. Endoscopic prediction of submucosal invasion in Barrett's cancer with the use of artificial intelligence: a pilot study. Endoscopy. 2021;53:878-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 24. | Ghatwary N, Zolgharni M, Ye X. Early esophageal adenocarcinoma detection using deep learning methods. Int J Comput Assist Radiol Surg. 2019;14:611-621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 25. | Iwagami H, Ishihara R, Aoyama K, Fukuda H, Shimamoto Y, Kono M, Nakahira H, Matsuura N, Shichijo S, Kanesaka T, Kanzaki H, Ishii T, Nakatani Y, Tada T. Artificial intelligence for the detection of esophageal and esophagogastric junctional adenocarcinoma. J Gastroenterol Hepatol. 2021;36:131-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 26. | Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598-5606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 736] [Cited by in RCA: 747] [Article Influence: 62.3] [Reference Citation Analysis (8)] |
| 27. | Shin D, Protano MA, Polydorides AD, Dawsey SM, Pierce MC, Kim MK, Schwarz RA, Quang T, Parikh N, Bhutani MS, Zhang F, Wang G, Xue L, Wang X, Xu H, Anandasabapathy S, Richards-Kortum RR. Quantitative analysis of high-resolution microendoscopic images for diagnosis of esophageal squamous cell carcinoma. Clin Gastroenterol Hepatol. 2015;13:272-279.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 28. | Quang T, Schwarz RA, Dawsey SM, Tan MC, Patel K, Yu X, Wang G, Zhang F, Xu H, Anandasabapathy S, Richards-Kortum R. A tablet-interfaced high-resolution microendoscope with automated image interpretation for real-time evaluation of esophageal squamous cell neoplasia. Gastrointest Endosc. 2016;84:834-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 29. | Cai SL, Li B, Tan WM, Niu XJ, Yu HH, Yao LQ, Zhou PH, Yan B, Zhong YS. Using a deep learning system in endoscopy for screening of early esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2019;90:745-753.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 30. | Liu G, Hua J, Wu Z, Meng T, Sun M, Huang P, He X, Sun W, Li X, Chen Y. Automatic classification of esophageal lesions in endoscopic images using a convolutional neural network. Ann Transl Med. 2020;8:486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 31. | Liu W, Yuan X, Guo L, Pan F, Wu C, Sun Z, Tian F, Yuan C, Zhang W, Bai S, Feng J, Hu Y, Hu B. Artificial Intelligence for Detecting and Delineating Margins of Early ESCC Under WLI Endoscopy. Clin Transl Gastroenterol. 2022;13:e00433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 32. | Ohmori M, Ishihara R, Aoyama K, Nakagawa K, Iwagami H, Matsuura N, Shichijo S, Yamamoto K, Nagaike K, Nakahara M, Inoue T, Aoi K, Okada H, Tada T. Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc. 2020;91:301-309.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 33. | Guo L, Xiao X, Wu C, Zeng X, Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, Wang X, Cheung O, Sharma M, Liu J, Hu B. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc. 2020;91:41-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 153] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 34. | Fukuda H, Ishihara R, Kato Y, Matsunaga T, Nishida T, Yamada T, Ogiyama H, Horie M, Kinoshita K, Tada T. Comparison of performances of artificial intelligence versus expert endoscopists for real-time assisted diagnosis of esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2020;92:848-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 35. | Li B, Cai SL, Tan WM, Li JC, Yalikong A, Feng XS, Yu HH, Lu PX, Feng Z, Yao LQ, Zhou PH, Yan B, Zhong YS. Comparative study on artificial intelligence systems for detecting early esophageal squamous cell carcinoma between narrow-band and white-light imaging. World J Gastroenterol. 2021;27:281-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 36. | Shiroma S, Yoshio T, Kato Y, Horie Y, Namikawa K, Tokai Y, Yoshimizu S, Yoshizawa N, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Akazawa N, Akiyama J, Tada T, Fujisaki J. Ability of artificial intelligence to detect T1 esophageal squamous cell carcinoma from endoscopic videos and the effects of real-time assistance. Sci Rep. 2021;11:7759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 37. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 272] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 38. | Kumagai Y, Takubo K, Kawada K, Aoyama K, Endo Y, Ozawa T, Hirasawa T, Yoshio T, Ishihara S, Fujishiro M, Tamaru JI, Mochiki E, Ishida H, Tada T. Diagnosis using deep-learning artificial intelligence based on the endocytoscopic observation of the esophagus. Esophagus. 2019;16:180-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 39. | Everson M, Herrera L, Li W, Luengo IM, Ahmad O, Banks M, Magee C, Alzoubaidi D, Hsu HM, Graham D, Vercauteren T, Lovat L, Ourselin S, Kashin S, Wang HP, Wang WL, Haidry RJ. Artificial intelligence for the real-time classification of intrapapillary capillary loop patterns in the endoscopic diagnosis of early oesophageal squamous cell carcinoma: A proof-of-concept study. United European Gastroenterol J. 2019;7:297-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 40. | Zhao YY, Xue DX, Wang YL, Zhang R, Sun B, Cai YP, Feng H, Cai Y, Xu JM. Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy. 2019;51:333-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 41. | Nakagawa K, Ishihara R, Aoyama K, Ohmori M, Nakahira H, Matsuura N, Shichijo S, Nishida T, Yamada T, Yamaguchi S, Ogiyama H, Egawa S, Kishida O, Tada T. Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest Endosc. 2019;90:407-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 42. | Shimamoto Y, Ishihara R, Kato Y, Shoji A, Inoue T, Matsueda K, Miyake M, Waki K, Kono M, Fukuda H, Matsuura N, Nagaike K, Aoi K, Yamamoto K, Nakahara M, Nishihara A, Tada T. Real-time assessment of video images for esophageal squamous cell carcinoma invasion depth using artificial intelligence. J Gastroenterol. 2020;55:1037-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 43. | Tokai Y, Yoshio T, Aoyama K, Horie Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Tsuchida T, Hirasawa T, Sakakibara Y, Yamada T, Yamaguchi S, Fujisaki J, Tada T. Application of artificial intelligence using convolutional neural networks in determining the invasion depth of esophageal squamous cell carcinoma. Esophagus. 2020;17:250-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 44. | Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, Ohnishi T, Fujishiro M, Matsuo K, Fujisaki J, Tada T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 426] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 45. | Lee JH, Kim YJ, Kim YW, Park S, Choi YI, Park DK, Kim KG, Chung JW. Spotting malignancies from gastric endoscopic images using deep learning. Surg Endosc. 2019;33:3790-3797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 46. | Li L, Chen Y, Shen Z, Zhang X, Sang J, Ding Y, Yang X, Li J, Chen M, Jin C, Chen C, Yu C. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer. 2020;23:126-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 137] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 47. | Miyaki R, Yoshida S, Tanaka S, Kominami Y, Sanomura Y, Matsuo T, Oka S, Raytchev B, Tamaki T, Koide T, Kaneda K, Yoshihara M, Chayama K. Quantitative identification of mucosal gastric cancer under magnifying endoscopy with flexible spectral imaging color enhancement. J Gastroenterol Hepatol. 2013;28:841-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 48. | Kanesaka T, Lee TC, Uedo N, Lin KP, Chen HZ, Lee JY, Wang HP, Chang HT. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest Endosc. 2018;87:1339-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 129] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 49. | Tang D, Wang L, Ling T, Lv Y, Ni M, Zhan Q, Fu Y, Zhuang D, Guo H, Dou X, Zhang W, Xu G, Zou X. Development and validation of a real-time artificial intelligence-assisted system for detecting early gastric cancer: A multicentre retrospective diagnostic study. EBioMedicine. 2020;62:103146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 50. | Wu L, Zhou W, Wan X, Zhang J, Shen L, Hu S, Ding Q, Mu G, Yin A, Huang X, Liu J, Jiang X, Wang Z, Deng Y, Liu M, Lin R, Ling T, Li P, Wu Q, Jin P, Chen J, Yu H. A deep neural network improves endoscopic detection of early gastric cancer without blind spots. Endoscopy. 2019;51:522-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 156] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 51. | Cho BJ, Bang CS, Park SW, Yang YJ, Seo SI, Lim H, Shin WG, Hong JT, Yoo YT, Hong SH, Choi JH, Lee JJ, Baik GH. Automated classification of gastric neoplasms in endoscopic images using a convolutional neural network. Endoscopy. 2019;51:1121-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (1)] |
| 52. | Namikawa K, Hirasawa T, Nakano K, Ikenoyama Y, Ishioka M, Shiroma S, Tokai Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Artificial intelligence-based diagnostic system classifying gastric cancers and ulcers: comparison between the original and newly developed systems. Endoscopy. 2020;52:1077-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 53. | Yuan XL, Zhou Y, Liu W, Luo Q, Zeng XH, Yi Z, Hu B. Artificial intelligence for diagnosing gastric lesions under white-light endoscopy. Surg Endosc. 2022;36:9444-9453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 54. | Guo L, Gong H, Wang Q, Zhang Q, Tong H, Li J, Lei X, Xiao X, Li C, Jiang J, Hu B, Song J, Tang C, Huang Z. Detection of multiple lesions of gastrointestinal tract for endoscopy using artificial intelligence model: a pilot study. Surg Endosc. 2021;35:6532-6538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 55. | Ikenoyama Y, Hirasawa T, Ishioka M, Namikawa K, Yoshimizu S, Horiuchi Y, Ishiyama A, Yoshio T, Tsuchida T, Takeuchi Y, Shichijo S, Katayama N, Fujisaki J, Tada T. Detecting early gastric cancer: Comparison between the diagnostic ability of convolutional neural networks and endoscopists. Dig Endosc. 2021;33:141-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 103] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 56. | Horiuchi Y, Aoyama K, Tokai Y, Hirasawa T, Yoshimizu S, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Convolutional Neural Network for Differentiating Gastric Cancer from Gastritis Using Magnified Endoscopy with Narrow Band Imaging. Dig Dis Sci. 2020;65:1355-1363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (1)] |
| 57. | Horiuchi Y, Hirasawa T, Ishizuka N, Tokai Y, Namikawa K, Yoshimizu S, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Performance of a computer-aided diagnosis system in diagnosing early gastric cancer using magnifying endoscopy videos with narrow-band imaging (with videos). Gastrointest Endosc. 2020;92:856-865.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 58. | Hu H, Gong L, Dong D, Zhu L, Wang M, He J, Shu L, Cai Y, Cai S, Su W, Zhong Y, Li C, Zhu Y, Fang M, Zhong L, Yang X, Zhou P, Tian J. Identifying early gastric cancer under magnifying narrow-band images with deep learning: a multicenter study. Gastrointest Endosc. 2021;93:1333-1341.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 59. | Ueyama H, Kato Y, Akazawa Y, Yatagai N, Komori H, Takeda T, Matsumoto K, Ueda K, Hojo M, Yao T, Nagahara A, Tada T. Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging. J Gastroenterol Hepatol. 2021;36:482-489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 89] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 60. | Yoon HJ, Kim S, Kim JH, Keum JS, Oh SI, Jo J, Chun J, Youn YH, Park H, Kwon IG, Choi SH, Noh SH. A Lesion-Based Convolutional Neural Network Improves Endoscopic Detection and Depth Prediction of Early Gastric Cancer. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 61. | Zhu Y, Wang QC, Xu MD, Zhang Z, Cheng J, Zhong YS, Zhang YQ, Chen WF, Yao LQ, Zhou PH, Li QL. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc. 2019;89:806-815.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 231] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 62. | Cho BJ, Bang CS, Lee JJ, Seo CW, Kim JH. Prediction of Submucosal Invasion for Gastric Neoplasms in Endoscopic Images Using Deep-Learning. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 63. | Nagao S, Tsuji Y, Sakaguchi Y, Takahashi Y, Minatsuki C, Niimi K, Yamashita H, Yamamichi N, Seto Y, Tada T, Koike K. Highly accurate artificial intelligence systems to predict the invasion depth of gastric cancer: efficacy of conventional white-light imaging, nonmagnifying narrow-band imaging, and indigo-carmine dye contrast imaging. Gastrointest Endosc. 2020;92:866-873.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 64. | Ku Y, Ding H, Wang G. Efficient Synchronous Real-Time CADe for Multicategory Lesions in Gastroscopy by Using Multiclass Detection Model. Biomed Res Int. 2022;2022:8504149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 65. | Watanabe K, Nagata N, Shimbo T, Nakashima R, Furuhata E, Sakurai T, Akazawa N, Yokoi C, Kobayakawa M, Akiyama J, Mizokami M, Uemura N. Accuracy of endoscopic diagnosis of Helicobacter pylori infection according to level of endoscopic experience and the effect of training. BMC Gastroenterol. 2013;13:128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 66. | Shichijo S, Nomura S, Aoyama K, Nishikawa Y, Miura M, Shinagawa T, Takiyama H, Tanimoto T, Ishihara S, Matsuo K, Tada T. Application of Convolutional Neural Networks in the Diagnosis of Helicobacter pylori Infection Based on Endoscopic Images. EBioMedicine. 2017;25:106-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 67. | Shichijo S, Endo Y, Aoyama K, Takeuchi Y, Ozawa T, Takiyama H, Matsuo K, Fujishiro M, Ishihara S, Ishihara R, Tada T. Application of convolutional neural networks for evaluating Helicobacter pylori infection status on the basis of endoscopic images. Scand J Gastroenterol. 2019;54:158-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 68. | Itoh T, Kawahira H, Nakashima H, Yata N. Deep learning analyzes Helicobacter pylori infection by upper gastrointestinal endoscopy images. Endosc Int Open. 2018;6:E139-E144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 130] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 69. | Zheng W, Zhang X, Kim JJ, Zhu X, Ye G, Ye B, Wang J, Luo S, Li J, Yu T, Liu J, Hu W, Si J. High Accuracy of Convolutional Neural Network for Evaluation of Helicobacter pylori Infection Based on Endoscopic Images: Preliminary Experience. Clin Transl Gastroenterol. 2019;10:e00109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 70. | Yasuda T, Hiroyasu T, Hiwa S, Okada Y, Hayashi S, Nakahata Y, Yasuda Y, Omatsu T, Obora A, Kojima T, Ichikawa H, Yagi N. Potential of automatic diagnosis system with linked color imaging for diagnosis of Helicobacter pylori infection. Dig Endosc. 2020;32:373-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 71. | Nakashima H, Kawahira H, Kawachi H, Sakaki N. Endoscopic three-categorical diagnosis of Helicobacter pylori infection using linked color imaging and deep learning: a single-center prospective study (with video). Gastric Cancer. 2020;23:1033-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 72. | Stoleru CA, Dulf EH, Ciobanu L. Automated detection of celiac disease using Machine Learning Algorithms. Sci Rep. 2022;12:4071. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 73. | Gadermayr M, Kogler H, Karla M, Merhof D, Uhl A, Vécsei A. Computer-aided texture analysis combined with experts' knowledge: Improving endoscopic celiac disease diagnosis. World J Gastroenterol. 2016;22:7124-7134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 74. | Wimmer G, Hegenbart S, Vecsei A, Uhl A. Convolutional Neural Network Architectures for the Automated Diagnosis of Celiac Disease. Presented at: International Workshop on Computer-Assisted and Robotic Endoscopy. CARE 2016; 104–113. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 75. | Zhou T, Han G, Li BN, Lin Z, Ciaccio EJ, Green PH, Qin J. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput Biol Med. 2017;85:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 76. | Wang X, Qian H, Ciaccio EJ, Lewis SK, Bhagat G, Green PH, Xu S, Huang L, Gao R, Liu Y. Celiac disease diagnosis from videocapsule endoscopy images with residual learning and deep feature extraction. Comput Methods Programs Biomed. 2020;187:105236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 77. | Yogapriya J, Chandran V, Sumithra MG, Anitha P, Jenopaul P, Suresh Gnana Dhas C. Gastrointestinal Tract Disease Classification from Wireless Endoscopy Images Using Pretrained Deep Learning Model. Comput Math Methods Med. 2021;2021:5940433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 78. | Charisis VS, Hadjileontiadis LJ, Liatsos CN, Mavrogiannis CC, Sergiadis GD. Capsule endoscopy image analysis using texture information from various colour models. Comput Methods Programs Biomed. 2012;107:61-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 79. | Charisis VS, Hadjileontiadis LJ. Potential of hybrid adaptive filtering in inflammatory lesion detection from capsule endoscopy images. World J Gastroenterol. 2016;22:8641-8657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 80. | Kumar R, Zhao Q, Seshamani S, Mullin G, Hager G, Dassopoulos T. Assessment of Crohn's disease lesions in wireless capsule endoscopy images. IEEE Trans Biomed Eng. 2012;59:355-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 81. | Wei Z, Wang W, Bradfield J, Li J, Cardinale C, Frackelton E, Kim C, Mentch F, Van Steen K, Visscher PM, Baldassano RN, Hakonarson H; International IBD Genetics Consortium. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease. Am J Hum Genet. 2013;92:1008-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 135] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 82. | Ferreira JPS, de Mascarenhas Saraiva MJDQEC, Afonso JPL, Ribeiro TFC, Cardoso HMC, Ribeiro Andrade AP, de Mascarenhas Saraiva MNG, Parente MPL, Natal Jorge R, Lopes SIO, de Macedo GMG. Identification of Ulcers and Erosions by the Novel Pillcam™ Crohn's Capsule Using a Convolutional Neural Network: A Multicentre Pilot Study. J Crohns Colitis. 2022;16:169-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 40] [Article Influence: 13.3] [Reference Citation Analysis (1)] |
| 83. | Klang E, Grinman A, Soffer S, Margalit Yehuda R, Barzilay O, Amitai MM, Konen E, Ben-Horin S, Eliakim R, Barash Y, Kopylov U. Automated Detection of Crohn's Disease Intestinal Strictures on Capsule Endoscopy Images Using Deep Neural Networks. J Crohns Colitis. 2021;15:749-756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 84. | Wu X, Chen H, Gan T, Chen J, Ngo CW, Peng Q. Automatic Hookworm Detection in Wireless Capsule Endoscopy Images. IEEE Trans Med Imaging. 2016;35:1741-1752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 85. | He JY, Wu X, Jiang YG, Peng Q, Jain R. Hookworm Detection in Wireless Capsule Endoscopy Images With Deep Learning. IEEE Trans Image Process. 2018;27:2379-2392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 86. | Gan T, Yang Y, Liu S, Zeng B, Yang J, Deng K, Wu J, Yang L. Automatic Detection of Small Intestinal Hookworms in Capsule Endoscopy Images Based on a Convolutional Neural Network. Gastroenterol Res Pract. 2021;2021:5682288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 87. | Sainju S, Bui FM, Wahid KA. Automated bleeding detection in capsule endoscopy videos using statistical features and region growing. J Med Syst. 2014;38:25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 88. | Usman MA, Satrya GB, Usman MR, Shin SY. Detection of small colon bleeding in wireless capsule endoscopy videos. Comput Med Imaging Graph. 2016;54:16-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 89. | Ghosh T, Chakareski J. Deep Transfer Learning for Automated Intestinal Bleeding Detection in Capsule Endoscopy Imaging. J Digit Imaging. 2021;34:404-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 90. | Tsuboi A, Oka S, Aoyama K, Saito H, Aoki T, Yamada A, Matsuda T, Fujishiro M, Ishihara S, Nakahori M, Koike K, Tanaka S, Tada T. Artificial intelligence using a convolutional neural network for automatic detection of small-bowel angioectasia in capsule endoscopy images. Dig Endosc. 2020;32:382-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 91. | Ribeiro T, Saraiva MM, Ferreira JPS, Cardoso H, Afonso J, Andrade P, Parente M, Jorge RN, Macedo G. Artificial intelligence and capsule endoscopy: automatic detection of vascular lesions using a convolutional neural network. Ann Gastroenterol. 2021;34:820-828. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 92. | Afonso J, Saraiva MM, Ferreira JPS, Cardoso H, Ribeiro T, Andrade P, Parente M, Jorge RN, Macedo G. Automated detection of ulcers and erosions in capsule endoscopy images using a convolutional neural network. Med Biol Eng Comput. 2022;60:719-725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 93. | Mascarenhas M, Ribeiro T, Afonso J, Ferreira JPS, Cardoso H, Andrade P, Parente MPL, Jorge RN, Mascarenhas Saraiva M, Macedo G. Deep learning and colon capsule endoscopy: automatic detection of blood and colonic mucosal lesions using a convolutional neural network. Endosc Int Open. 2022;10:E171-E177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 94. | Lewis BS, Eisen GM, Friedman S. A pooled analysis to evaluate results of capsule endoscopy trials. Endoscopy. 2005;37:960-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 187] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 95. | Li B, Meng MQ. Tumor recognition in wireless capsule endoscopy images using textural features and SVM-based feature selection. IEEE Trans Inf Technol Biomed. 2012;16:323-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 96. | Liu G, Yan G, Kuang S, Wang Y. Detection of small bowel tumor based on multi-scale curvelet analysis and fractal technology in capsule endoscopy. Comput Biol Med. 2016;70:131-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 97. | Faghih Dinevari V, Karimian Khosroshahi G, Zolfy Lighvan M. Singular Value Decomposition Based Features for Automatic Tumor Detection in Wireless Capsule Endoscopy Images. Appl Bionics Biomech. 2016;2016:3678913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 98. | Shanmuga Sundaram P, Santhiyakumari N. An Enhancement of Computer Aided Approach for Colon Cancer Detection in WCE Images Using ROI Based Color Histogram and SVM2. J Med Syst. 2019;43:29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 99. | Blanes-Vidal V, Baatrup G, Nadimi ES. Addressing priority challenges in the detection and assessment of colorectal polyps from capsule endoscopy and colonoscopy in colorectal cancer screening using machine learning. Acta Oncol. 2019;58:S29-S36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 100. | Saraiva MM, Ferreira JPS, Cardoso H, Afonso J, Ribeiro T, Andrade P, Parente MPL, Jorge RN, Macedo G. Artificial intelligence and colon capsule endoscopy: development of an automated diagnostic system of protruding lesions in colon capsule endoscopy. Tech Coloproctol. 2021;25:1243-1248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (1)] |
| 101. | Mascarenhas M, Afonso J, Ribeiro T, Cardoso H, Andrade P, Ferreira JPS, Saraiva MM, Macedo G. Performance of a Deep Learning System for Automatic Diagnosis of Protruding Lesions in Colon Capsule Endoscopy. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 102. | Constantinescu AF, Ionescu M, Iovănescu VF, Ciurea ME, Ionescu AG, Streba CT, Bunescu MG, Rogoveanu I, Vere CC. A computer-aided diagnostic system for intestinal polyps identified by wireless capsule endoscopy. Rom J Morphol Embryol. 2016;57:979-984. [PubMed] |
| 103. | Xia J, Xia T, Pan J, Gao F, Wang S, Qian YY, Wang H, Zhao J, Jiang X, Zou WB, Wang YC, Zhou W, Li ZS, Liao Z. Use of artificial intelligence for detection of gastric lesions by magnetically controlled capsule endoscopy. Gastrointest Endosc. 2021;93:133-139.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 104. | Yuan Y, Meng MQ. Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys. 2017;44:1379-1389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 105. | Sharma P, Burke CA, Johnson DA, Cash BD. The importance of colonoscopy bowel preparation for the detection of colorectal lesions and colorectal cancer prevention. Endosc Int Open. 2020;8:E673-E683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 106. | Ahmad OF. Deep learning for automated bowel preparation assessment during colonoscopy: time to embrace a new approach? Lancet Digit Health. 2021;3:e685-e686. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 107. | Hassan C, East J, Radaelli F, Spada C, Benamouzig R, Bisschops R, Bretthauer M, Dekker E, Dinis-Ribeiro M, Ferlitsch M, Fuccio L, Awadie H, Gralnek I, Jover R, Kaminski MF, Pellisé M, Triantafyllou K, Vanella G, Mangas-Sanjuan C, Frazzoni L, Van Hooft JE, Dumonceau JM. Bowel preparation for colonoscopy: European Society of Gastrointestinal Endoscopy (ESGE) Guideline - Update 2019. Endoscopy. 2019;51:775-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 351] [Article Influence: 58.5] [Reference Citation Analysis (4)] |
| 108. | Rex DK, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T, Levin TR, Lieberman D, Robertson DJ. Colorectal Cancer Screening: Recommendations for Physicians and Patients from the U.S. Multi-Society Task Force on Colorectal Cancer. Am J Gastroenterol. 2017;112:1016-1030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 475] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 109. | ASGE Standards of Practice Committee; Saltzman JR, Cash BD, Pasha SF, Early DS, Muthusamy VR, Khashab MA, Chathadi KV, Fanelli RD, Chandrasekhara V, Lightdale JR, Fonkalsrud L, Shergill AK, Hwang JH, Decker GA, Jue TL, Sharaf R, Fisher DA, Evans JA, Foley K, Shaukat A, Eloubeidi MA, Faulx AL, Wang A, Acosta RD. Bowel preparation before colonoscopy. Gastrointest Endosc. 2015;81:781-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 305] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 110. | Hadlock SD, Liu N, Bernstein M, Gould M, Rabeneck L, Ruco A, Sutradhar R, Tinmouth JM. The Quality of Colonoscopy Reporting in Usual Practice: Are Endoscopists Reporting Key Data Elements? Can J Gastroenterol Hepatol. 2016;2016:1929361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 111. | Gavin DR, Valori RM, Anderson JT, Donnelly MT, Williams JG, Swarbrick ET. The national colonoscopy audit: a nationwide assessment of the quality and safety of colonoscopy in the UK. Gut. 2013;62:242-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 207] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 112. | Coe SG, Panjala C, Heckman MG, Patel M, Qumseya BJ, Wang YR, Dalton B, Tran P, Palmer W, Diehl N, Wallace MB, Raimondo M. Quality in colonoscopy reporting: an assessment of compliance and performance improvement. Dig Liver Dis. 2012;44:660-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 113. | Zhou J, Wu L, Wan X, Shen L, Liu J, Zhang J, Jiang X, Wang Z, Yu S, Kang J, Li M, Hu S, Hu X, Gong D, Chen D, Yao L, Zhu Y, Yu H. A novel artificial intelligence system for the assessment of bowel preparation (with video). Gastrointest Endosc. 2020;91:428-435.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 114. | Zhou W, Yao L, Wu H, Zheng B, Hu S, Zhang L, Li X, He C, Wang Z, Li Y, Huang C, Guo M, Zhang X, Zhu Q, Wu L, Deng Y, Zhang J, Tan W, Li C, Zhang C, Gong R, Du H, Zhou J, Sharma P, Yu H. Multi-step validation of a deep learning-based system for the quantification of bowel preparation: a prospective, observational study. Lancet Digit Health. 2021;3:e697-e706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 115. | Lai EJ, Calderwood AH, Doros G, Fix OK, Jacobson BC. The Boston bowel preparation scale: a valid and reliable instrument for colonoscopy-oriented research. Gastrointest Endosc. 2009;69:620-625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 930] [Cited by in RCA: 924] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 116. | Lee JY, Calderwood AH, Karnes W, Requa J, Jacobson BC, Wallace MB. Artificial intelligence for the assessment of bowel preparation. Gastrointest Endosc. 2022;95:512-518.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 117. | Low DJ, Hong Z, Jugnundan S, Mukherjee A, Grover SC. Automated Detection of Bowel Preparation Scoring and Adequacy With Deep Convolutional Neural Networks. JCAG. 2022;XX:1-5. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 118. | Wang YP, Jheng YC, Sung KY, Lin HE, Hsin IF, Chen PH, Chu YC, Lu D, Wang YJ, Hou MC, Lee FY, Lu CL. Use of U-Net Convolutional Neural Networks for Automated Segmentation of Fecal Material for Objective Evaluation of Bowel Preparation Quality in Colonoscopy. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 119. | Kohli A, Holzwanger EA, Levy AN. Emerging use of artificial intelligence in inflammatory bowel disease. World J Gastroenterol. 2020;26:6923-6928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 120. | Cohen-Mekelburg S, Berry S, Stidham RW, Zhu J, Waljee AK. Clinical applications of artificial intelligence and machine learning-based methods in inflammatory bowel disease. J Gastroenterol Hepatol. 2021;36:279-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 121. | Ozawa T, Ishihara S, Fujishiro M, Saito H, Kumagai Y, Shichijo S, Aoyama K, Tada T. Novel computer-assisted diagnosis system for endoscopic disease activity in patients with ulcerative colitis. Gastrointest Endosc. 2019;89:416-421.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 147] [Article Influence: 24.5] [Reference Citation Analysis (1)] |
| 122. | Stidham RW, Liu W, Bishu S, Rice MD, Higgins PDR, Zhu J, Nallamothu BK, Waljee AK. Performance of a Deep Learning Model vs Human Reviewers in Grading Endoscopic Disease Severity of Patients With Ulcerative Colitis. JAMA Netw Open. 2019;2:e193963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 174] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 123. | Dave M, Loftus EV Jr. Mucosal healing in inflammatory bowel disease-a true paradigm of success? Gastroenterol Hepatol (N Y). 2012;8:29-38. [PubMed] |
| 124. | Huang TY, Zhan SQ, Chen PJ, Yang CW, Lu HH. Accurate diagnosis of endoscopic mucosal healing in ulcerative colitis using deep learning and machine learning. J Chin Med Assoc. 2021;84:678-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 125. | Barreiro-de Acosta M, Vallejo N, de la Iglesia D, Uribarri L, Bastón I, Ferreiro-Iglesias R, Lorenzo A, Domínguez-Muñoz JE. Evaluation of the Risk of Relapse in Ulcerative Colitis According to the Degree of Mucosal Healing (Mayo 0 vs 1): A Longitudinal Cohort Study. J Crohns Colitis. 2016;10:13-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 178] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 126. | Osterman MT, Scott FI, Fogt FF, Gilroy ED, Parrott S, Galanko J, Cross R, Moss A, Herfarth HH, Higgins PDR. Endoscopic and Histological Assessment, Correlation, and Relapse in Clinically Quiescent Ulcerative Colitis (MARQUEE). Inflamm Bowel Dis. 2021;27:207-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 127. | Bhambhvani HP, Zamora A. Deep learning enabled classification of Mayo endoscopic subscore in patients with ulcerative colitis. Eur J Gastroenterol Hepatol. 2021;33:645-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 128. | Yao H, Najarian K, Gryak J, Bishu S, Rice MD, Waljee AK, Wilkins HJ, Stidham RW. Fully automated endoscopic disease activity assessment in ulcerative colitis. Gastrointest Endosc. 2021;93:728-736.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 129. | Gottlieb K, Requa J, Karnes W, Chandra Gudivada R, Shen J, Rael E, Arora V, Dao T, Ninh A, McGill J. Central Reading of Ulcerative Colitis Clinical Trial Videos Using Neural Networks. Gastroenterology. 2021;160:710-719.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 96] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 130. | Gutierrez Becker B, Arcadu F, Thalhammer A, Gamez Serna C, Feehan O, Drawnel F, Oh YS, Prunotto M. Training and deploying a deep learning model for endoscopic severity grading in ulcerative colitis using multicenter clinical trial data. Ther Adv Gastrointest Endosc. 2021;14:2631774521990623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 131. | Maeda Y, Kudo SE, Mori Y, Misawa M, Ogata N, Sasanuma S, Wakamura K, Oda M, Mori K, Ohtsuka K. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc. 2019;89:408-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 164] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 132. | Takenaka K, Ohtsuka K, Fujii T, Negi M, Suzuki K, Shimizu H, Oshima S, Akiyama S, Motobayashi M, Nagahori M, Saito E, Matsuoka K, Watanabe M. Development and Validation of a Deep Neural Network for Accurate Evaluation of Endoscopic Images From Patients With Ulcerative Colitis. Gastroenterology. 2020;158:2150-2157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 180] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 133. | Takenaka K, Ohtsuka K, Fujii T, Oshima S, Okamoto R, Watanabe M. Deep Neural Network Accurately Predicts Prognosis of Ulcerative Colitis Using Endoscopic Images. Gastroenterology. 2021;160:2175-2177.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 134. | Takenaka K, Fujii T, Kawamoto A, Suzuki K, Shimizu H, Maeyashiki C, Yamaji O, Motobayashi M, Igarashi A, Hanazawa R, Hibiya S, Nagahori M, Saito E, Okamoto R, Ohtsuka K, Watanabe M. Deep neural network for video colonoscopy of ulcerative colitis: a cross-sectional study. Lancet Gastroenterol Hepatol. 2022;7:230-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 135. | Udristoiu AL, Stefanescu D, Gruionu G, Gruionu LG, Iacob AV, Karstensen JG, Vilman P, Saftoiu A. Deep Learning Algorithm for the Confirmation of Mucosal Healing in Crohn's Disease, Based on Confocal Laser Endomicroscopy Images. J Gastrointestin Liver Dis. 2021;30:59-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 136. | Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14:101174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 687] [Cited by in RCA: 1344] [Article Influence: 336.0] [Reference Citation Analysis (5)] |
| 137. | Roselló S, Simón S, Cervantes A. Programmed colorectal cancer screening decreases incidence and mortality. Transl Gastroenterol Hepatol. 2019;4:84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 138. | Ahn SB, Han DS, Bae JH, Byun TJ, Kim JP, Eun CS. The Miss Rate for Colorectal Adenoma Determined by Quality-Adjusted, Back-to-Back Colonoscopies. Gut Liver. 2012;6:64-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 142] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 139. | Komeda Y, Handa H, Watanabe T, Nomura T, Kitahashi M, Sakurai T, Okamoto A, Minami T, Kono M, Arizumi T, Takenaka M, Hagiwara S, Matsui S, Nishida N, Kashida H, Kudo M. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology. 2017;93 Suppl 1:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 140. | Misawa M, Kudo SE, Mori Y, Cho T, Kataoka S, Yamauchi A, Ogawa Y, Maeda Y, Takeda K, Ichimasa K, Nakamura H, Yagawa Y, Toyoshima N, Ogata N, Kudo T, Hisayuki T, Hayashi T, Wakamura K, Baba T, Ishida F, Itoh H, Roth H, Oda M, Mori K. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology. 2018;154:2027-2029.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 269] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 141. | Hassan C, Wallace MB, Sharma P, Maselli R, Craviotto V, Spadaccini M, Repici A. New artificial intelligence system: first validation study versus experienced endoscopists for colorectal polyp detection. Gut. 2020;69:799-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 123] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 142. | Guo Z, Nemoto D, Zhu X, Li Q, Aizawa M, Utano K, Isohata N, Endo S, Kawarai Lefor A, Togashi K. Polyp detection algorithm can detect small polyps: Ex vivo reading test compared with endoscopists. Dig Endosc. 2021;33:162-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 143. | Neumann H, Kreft A, Sivanathan V, Rahman F, Galle PR. Evaluation of novel LCI CAD EYE system for real time detection of colon polyps. PLoS One. 2021;16:e0255955. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 144. | Li JW, Chia T, Fock KM, Chong KW, Wong YJ, Ang TL. Artificial intelligence and polyp detection in colonoscopy: Use of a single neural network to achieve rapid polyp localization for clinical use. J Gastroenterol Hepatol. 2021;36:3298-3307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 145. | Ahmad OF, González-Bueno Puyal J, Brandao P, Kader R, Abbasi F, Hussein M, Haidry RJ, Toth D, Mountney P, Seward E, Vega R, Stoyanov D, Lovat LB. Performance of artificial intelligence for detection of subtle and advanced colorectal neoplasia. Dig Endosc. 2022;34:862-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (1)] |
| 146. | Hori K, Ikematsu H, Yamamoto Y, Matsuzaki H, Takeshita N, Shinmura K, Yoda Y, Kiuchi T, Takemoto S, Yokota H, Yano T. Detecting colon polyps in endoscopic images using artificial intelligence constructed with automated collection of annotated images from an endoscopy reporting system. Dig Endosc. 2022;34:1021-1029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 147. | Brand M, Troya J, Krenzer A, Saßmannshausen Z, Zoller WG, Meining A, Lux TJ, Hann A. Development and evaluation of a deep learning model to improve the usability of polyp detection systems during interventions. United European Gastroenterol J. 2022;10:477-484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 148. | Lux TJ, Banck M, Saßmannshausen Z, Troya J, Krenzer A, Fitting D, Sudarevic B, Zoller WG, Puppe F, Meining A, Hann A. Pilot study of a new freely available computer-aided polyp detection system in clinical practice. Int J Colorectal Dis. 2022;37:1349-1354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 149. | Wang P, Xiao X, Glissen Brown JR, Berzin TM, Tu M, Xiong F, Hu X, Liu P, Song Y, Zhang D, Yang X, Li L, He J, Yi X, Liu J, Liu X. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng. 2018;2:741-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 270] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 150. | Shen P, Li WZ, Li JX, Pei ZC, Luo YX, Mu JB, Li W, Wang XM. Real-time use of a computer-aided system for polyp detection during colonoscopy, an ambispective study. J Dig Dis. 2021;22:256-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 151. | Livovsky DM, Veikherman D, Golany T, Aides A, Dashinsky V, Rabani N, Ben Shimol D, Blau Y, Katzir L, Shimshoni I, Liu Y, Segol O, Goldin E, Corrado G, Lachter J, Matias Y, Rivlin E, Freedman D. Detection of elusive polyps using a large-scale artificial intelligence system (with videos). Gastrointest Endosc. 2021;94:1099-1109.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 152. | Pacal I, Karaman A, Karaboga D, Akay B, Basturk A, Nalbantoglu U, Coskun S. An efficient real-time colonic polyp detection with YOLO algorithms trained by using negative samples and large datasets. Comput Biol Med. 2022;141:105031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 153. | Quan SY, Wei MT, Lee J, Mohi-Ud-Din R, Mostaghim R, Sachdev R, Siegel D, Friedlander Y, Friedland S. Clinical evaluation of a real-time artificial intelligence-based polyp detection system: a US multi-center pilot study. Sci Rep. 2022;12:6598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 154. | Horiuchi H, Tamai N, Kamba S, Inomata H, Ohya TR, Sumiyama K. Real-time computer-aided diagnosis of diminutive rectosigmoid polyps using an auto-fluorescence imaging system and novel color intensity analysis software. Scand J Gastroenterol. 2019;54:800-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 155. | Becq A, Chandnani M, Bharadwaj S, Baran B, Ernest-Suarez K, Gabr M, Glissen-Brown J, Sawhney M, Pleskow DK, Berzin TM. Effectiveness of a Deep-learning Polyp Detection System in Prospectively Collected Colonoscopy Videos With Variable Bowel Preparation Quality. J Clin Gastroenterol. 2020;54:554-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 156. | Wang P, Liu P, Glissen Brown JR, Berzin TM, Zhou G, Lei S, Liu X, Li L, Xiao X. Lower Adenoma Miss Rate of Computer-Aided Detection-Assisted Colonoscopy vs Routine White-Light Colonoscopy in a Prospective Tandem Study. Gastroenterology. 2020;159:1252-1261.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 147] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 157. | Shaukat A, Colucci D, Erisson L, Phillips S, Ng J, Iglesias JE, Saltzman JR, Somers S, Brugge W. Improvement in adenoma detection using a novel artificial intelligence-aided polyp detection device. Endosc Int Open. 2021;9:E263-E270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 158. | Pfeifer L, Neufert C, Leppkes M, Waldner MJ, Häfner M, Beyer A, Hoffman A, Siersema PD, Neurath MF, Rath T. Computer-aided detection of colorectal polyps using a newly generated deep convolutional neural network: from development to first clinical experience. Eur J Gastroenterol Hepatol. 2021;33:e662-e669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 159. | Ishiyama M, Kudo SE, Misawa M, Mori Y, Maeda Y, Ichimasa K, Kudo T, Hayashi T, Wakamura K, Miyachi H, Ishida F, Itoh H, Oda M, Mori K. Impact of the clinical use of artificial intelligence-assisted neoplasia detection for colonoscopy: a large-scale prospective, propensity score-matched study (with video). Gastrointest Endosc. 2022;95:155-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 160. | Zippelius C, Alqahtani SA, Schedel J, Brookman-Amissah D, Muehlenberg K, Federle C, Salzberger A, Schorr W, Pech O. Diagnostic accuracy of a novel artificial intelligence system for adenoma detection in daily practice: a prospective nonrandomized comparative study. Endoscopy. 2022;54:465-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 161. | Wu L, Zhang J, Zhou W, An P, Shen L, Liu J, Jiang X, Huang X, Mu G, Wan X, Lv X, Gao J, Cui N, Hu S, Chen Y, Hu X, Li J, Chen D, Gong D, He X, Ding Q, Zhu X, Li S, Wei X, Li X, Wang X, Zhou J, Zhang M, Yu HG. Randomised controlled trial of WISENSE, a real-time quality improving system for monitoring blind spots during esophagogastroduodenoscopy. Gut. 2019;68:2161-2169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 214] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 162. | Shaukat A, Lichtenstein DR, Somers SC, Chung DC, Perdue DG, Gopal M, Colucci DR, Phillips SA, Marka NA, Church TR, Brugge WR; SKOUT™ Registration Study Team. Computer-Aided Detection Improves Adenomas per Colonoscopy for Screening and Surveillance Colonoscopy: A Randomized Trial. Gastroenterology. 2022;163:732-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 75] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 163. | Glissen Brown JR, Mansour NM, Wang P, Chuchuca MA, Minchenberg SB, Chandnani M, Liu L, Gross SA, Sengupta N, Berzin TM. Deep Learning Computer-aided Polyp Detection Reduces Adenoma Miss Rate: A United States Multi-center Randomized Tandem Colonoscopy Study (CADeT-CS Trial). Clin Gastroenterol Hepatol. 2022;20:1499-1507.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 114] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 164. | Kamba S, Tamai N, Saitoh I, Matsui H, Horiuchi H, Kobayashi M, Sakamoto T, Ego M, Fukuda A, Tonouchi A, Shimahara Y, Nishikawa M, Nishino H, Saito Y, Sumiyama K. Reducing adenoma miss rate of colonoscopy assisted by artificial intelligence: a multicenter randomized controlled trial. J Gastroenterol. 2021;56:746-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 165. | Repici A, Spadaccini M, Antonelli G, Correale L, Maselli R, Galtieri PA, Pellegatta G, Capogreco A, Milluzzo SM, Lollo G, Di Paolo D, Badalamenti M, Ferrara E, Fugazza A, Carrara S, Anderloni A, Rondonotti E, Amato A, De Gottardi A, Spada C, Radaelli F, Savevski V, Wallace MB, Sharma P, Rösch T, Hassan C. Artificial intelligence and colonoscopy experience: lessons from two randomised trials. Gut. 2022;71:757-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 134] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 166. | Gong D, Wu L, Zhang J, Mu G, Shen L, Liu J, Wang Z, Zhou W, An P, Huang X, Jiang X, Li Y, Wan X, Hu S, Chen Y, Hu X, Xu Y, Zhu X, Li S, Yao L, He X, Chen D, Huang L, Wei X, Wang X, Yu H. Detection of colorectal adenomas with a real-time computer-aided system (ENDOANGEL): a randomised controlled study. Lancet Gastroenterol Hepatol. 2020;5:352-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 265] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 167. | Wang P, Liu X, Berzin TM, Glissen Brown JR, Liu P, Zhou C, Lei L, Li L, Guo Z, Lei S, Xiong F, Wang H, Song Y, Pan Y, Zhou G. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol. 2020;5:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 294] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 168. | Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, Li Y, Xu G, Tu M, Liu X. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 547] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 169. | Su JR, Li Z, Shao XJ, Ji CR, Ji R, Zhou RC, Li GC, Liu GQ, He YS, Zuo XL, Li YQ. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: a prospective randomized controlled study (with videos). Gastrointest Endosc. 2020;91:415-424.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 208] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 170. | Repici A, Badalamenti M, Maselli R, Correale L, Radaelli F, Rondonotti E, Ferrara E, Spadaccini M, Alkandari A, Fugazza A, Anderloni A, Galtieri PA, Pellegatta G, Carrara S, Di Leo M, Craviotto V, Lamonaca L, Lorenzetti R, Andrealli A, Antonelli G, Wallace M, Sharma P, Rosch T, Hassan C. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology. 2020;159:512-520.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 393] [Article Influence: 78.6] [Reference Citation Analysis (0)] |
| 171. | Liu P, Wang P, Glissen Brown JR, Berzin TM, Zhou G, Liu W, Xiao X, Chen Z, Zhang Z, Zhou C, Lei L, Xiong F, Li L, Liu X. The single-monitor trial: an embedded CADe system increased adenoma detection during colonoscopy: a prospective randomized study. Therap Adv Gastroenterol. 2020;13:1756284820979165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 172. | Xu L, He X, Zhou J, Zhang J, Mao X, Ye G, Chen Q, Xu F, Sang J, Wang J, Ding Y, Li Y, Yu C. Artificial intelligence-assisted colonoscopy: A prospective, multicenter, randomized controlled trial of polyp detection. Cancer Med. 2021;10:7184-7193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 173. | Liu WN, Zhang YY, Bian XQ, Wang LJ, Yang Q, Zhang XD, Huang J. Study on detection rate of polyps and adenomas in artificial-intelligence-aided colonoscopy. Saudi J Gastroenterol. 2020;26:13-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 126] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 174. | Luo Y, Zhang Y, Liu M, Lai Y, Liu P, Wang Z, Xing T, Huang Y, Li Y, Li A, Wang Y, Luo X, Liu S, Han Z. Artificial Intelligence-Assisted Colonoscopy for Detection of Colon Polyps: a Prospective, Randomized Cohort Study. J Gastrointest Surg. 2021;25:2011-2018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 175. | Barua I, Vinsard DG, Jodal HC, Løberg M, Kalager M, Holme Ø, Misawa M, Bretthauer M, Mori Y. Artificial intelligence for polyp detection during colonoscopy: a systematic review and meta-analysis. Endoscopy. 2021;53:277-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 158] [Article Influence: 39.5] [Reference Citation Analysis (1)] |
| 176. | Aziz M, Fatima R, Dong C, Lee-Smith W, Nawras A. The impact of deep convolutional neural network-based artificial intelligence on colonoscopy outcomes: A systematic review with meta-analysis. J Gastroenterol Hepatol. 2020;35:1676-1683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 177. | Hassan C, Spadaccini M, Iannone A, Maselli R, Jovani M, Chandrasekar VT, Antonelli G, Yu H, Areia M, Dinis-Ribeiro M, Bhandari P, Sharma P, Rex DK, Rösch T, Wallace M, Repici A. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastrointest Endosc. 2021;93:77-85.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 307] [Article Influence: 76.8] [Reference Citation Analysis (1)] |
| 178. | Li J, Lu J, Yan J, Tan Y, Liu D. Artificial intelligence can increase the detection rate of colorectal polyps and adenomas: a systematic review and meta-analysis. Eur J Gastroenterol Hepatol. 2021;33:1041-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 179. | Zhang Y, Zhang X, Wu Q, Gu C, Wang Z. Artificial Intelligence-Aided Colonoscopy for Polyp Detection: A Systematic Review and Meta-Analysis of Randomized Clinical Trials. J Laparoendosc Adv Surg Tech A. 2021;31:1143-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (1)] |
| 180. | Deliwala SS, Hamid K, Barbarawi M, Lakshman H, Zayed Y, Kandel P, Malladi S, Singh A, Bachuwa G, Gurvits GE, Chawla S. Artificial intelligence (AI) real-time detection vs. routine colonoscopy for colorectal neoplasia: a meta-analysis and trial sequential analysis. Int J Colorectal Dis. 2021;36:2291-2303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (1)] |
| 181. | Nazarian S, Glover B, Ashrafian H, Darzi A, Teare J. Diagnostic Accuracy of Artificial Intelligence and Computer-Aided Diagnosis for the Detection and Characterization of Colorectal Polyps: Systematic Review and Meta-analysis. J Med Internet Res. 2021;23:e27370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 182. | Xu Y, Ding W, Wang Y, Tan Y, Xi C, Ye N, Wu D, Xu X. Comparison of diagnostic performance between convolutional neural networks and human endoscopists for diagnosis of colorectal polyp: A systematic review and meta-analysis. PLoS One. 2021;16:e0246892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 183. | Bang CS, Lee JJ, Baik GH. Computer-Aided Diagnosis of Diminutive Colorectal Polyps in Endoscopic Images: Systematic Review and Meta-analysis of Diagnostic Test Accuracy. J Med Internet Res. 2021;23:e29682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 184. | Yoon D, Kong HJ, Kim BS, Cho WS, Lee JC, Cho M, Lim MH, Yang SY, Lim SH, Lee J, Song JH, Chung GE, Choi JM, Kang HY, Bae JH, Kim S. Colonoscopic image synthesis with generative adversarial network for enhanced detection of sessile serrated lesions using convolutional neural network. Sci Rep. 2022;12:261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 185. | Nemoto D, Guo Z, Peng B, Zhang R, Nakajima Y, Hayashi Y, Yamashina T, Aizawa M, Utano K, Lefor AK, Zhu X, Togashi K. Computer-aided diagnosis of serrated colorectal lesions using non-magnified white-light endoscopic images. Int J Colorectal Dis. 2022;37:1875-1884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 186. | Yao L, Zhang J, Liu J, Zhu L, Ding X, Chen D, Wu H, Lu Z, Zhou W, Zhang L, Xu B, Hu S, Zheng B, Yang Y, Yu H. A deep learning-based system for bile duct annotation and station recognition in linear endoscopic ultrasound. EBioMedicine. 2021;65:103238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 187. | Săftoiu A, Vilmann P, Gorunescu F, Janssen J, Hocke M, Larsen M, Iglesias-Garcia J, Arcidiacono P, Will U, Giovannini M, Dietrich CF, Havre R, Gheorghe C, McKay C, Gheonea DI, Ciurea T; European EUS Elastography Multicentric Study Group. Efficacy of an artificial neural network-based approach to endoscopic ultrasound elastography in diagnosis of focal pancreatic masses. Clin Gastroenterol Hepatol. 2012;10:84-90.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 128] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 188. | Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, Obata M, Kurita Y, Koda H, Toriyama K, Onishi S, Ishihara M, Tanaka T, Tajika M, Niwa Y. Usefulness of Deep Learning Analysis for the Diagnosis of Malignancy in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin Transl Gastroenterol. 2019;10:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 127] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 189. | Goyal H, Sherazi SAA, Gupta S, Perisetti A, Achebe I, Ali A, Tharian B, Thosani N, Sharma NR. Application of artificial intelligence in diagnosis of pancreatic malignancies by endoscopic ultrasound: a systemic review. Therap Adv Gastroenterol. 2022;15:17562848221093873. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 190. | Dumitrescu EA, Ungureanu BS, Cazacu IM, Florescu LM, Streba L, Croitoru VM, Sur D, Croitoru A, Turcu-Stiolica A, Lungulescu CV. Diagnostic Value of Artificial Intelligence-Assisted Endoscopic Ultrasound for Pancreatic Cancer: A Systematic Review and Meta-Analysis. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 191. | Minoda Y, Ihara E, Komori K, Ogino H, Otsuka Y, Chinen T, Tsuda Y, Ando K, Yamamoto H, Ogawa Y. Efficacy of endoscopic ultrasound with artificial intelligence for the diagnosis of gastrointestinal stromal tumors. J Gastroenterol. 2020;55:1119-1126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 192. | Jang SI, Kim YJ, Kim EJ, Kang H, Shon SJ, Seol YJ, Lee DK, Kim KG, Cho JH. Diagnostic performance of endoscopic ultrasound-artificial intelligence using deep learning analysis of gallbladder polypoid lesions. J Gastroenterol Hepatol. 2021;36:3548-3555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 193. | Zhang B, Zhu F, Li P, Zhu J. Artificial intelligence-assisted endoscopic ultrasound in the diagnosis of gastrointestinal stromal tumors: a meta-analysis. Surg Endosc. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 194. | Ye XH, Zhao LL, Wang L. Diagnostic accuracy of endoscopic ultrasound with artificial intelligence for gastrointestinal stromal tumors: A meta-analysis. J Dig Dis. 2022;23:253-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 195. | Marya NB, Powers PD, Petersen BT, Law R, Storm A, Abusaleh RR, Rau P, Stead C, Levy MJ, Martin J, Vargas EJ, Abu Dayyeh BK, Chandrasekhara V. Identification of patients with malignant biliary strictures using a cholangioscopy-based deep learning artificial intelligence (with video). Gastrointest Endosc. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 42] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 196. | Huang L, Lu X, Huang X, Zou X, Wu L, Zhou Z, Wu D, Tang D, Chen D, Wan X, Zhu Z, Deng T, Shen L, Liu J, Zhu Y, Gong D, Zhong Y, Liu F, Yu H. Intelligent difficulty scoring and assistance system for endoscopic extraction of common bile duct stones based on deep learning: multicenter study. Endoscopy. 2021;53:491-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 197. | Uche-Anya E, Anyane-Yeboa A, Berzin TM, Ghassemi M, May FP. Artificial intelligence in gastroenterology and hepatology: how to advance clinical practice while ensuring health equity. Gut. 2022;71:1909-1915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 63] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 198. | Stidham RW. Artificial Intelligence for Understanding Imaging, Text, and Data in Gastroenterology. Gastroenterol Hepatol (N Y). 2020;16:341-349. [PubMed] |
| 199. | Spadaccini M, Marco A, Franchellucci G, Sharma P, Hassan C, Repici A. Discovering the first US FDA-approved computer-aided polyp detection system. Future Oncol. 2022;18:1405-1412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |