Published online Dec 29, 2021. doi: 10.35713/aic.v2.i6.79
Peer-review started: December 9, 2021
First decision: December 13, 2021
Revised: December 22, 2021
Accepted: December 28, 2021
Article in press: December 28, 2021
Published online: December 29, 2021
Processing time: 19 Days and 22.3 Hours
Artificial intelligence (AI) is a new branch of computer science involving many disciplines and technologies. Since its application in the medical field, it has been constantly studied and developed. AI includes machine learning and neural networks to create new technologies or to improve existing ones. Various AI supporting systems are available for a personalized and novel strategy for the management of colorectal cancer (CRC). This mini-review aims to summarize the progress of research and possible clinical applications of AI in the investigation, early diagnosis, treatment, and management of CRC, to offer elements of knowledge as a starting point for new studies and future applications.
Core Tip: Many authors have summarized artificial intelligence applications in the field of cancer. This mini-review intends to open a window on the attempts being made on the application of artificial intelligence in the scientific and clinical research of colorectal cancer by summarizing the most evident results. Our aim is not to draw definitive conclusions but to stimulate the interest of researchers in the application of these new technologies, which seem to be able to offer valuable help in the near future.
- Citation: Cianci P, Restini E. Artificial intelligence in colorectal cancer management. Artif Intell Cancer 2021; 2(6): 79-89
- URL: https://www.wjgnet.com/2644-3228/full/v2/i6/79.htm
- DOI: https://dx.doi.org/10.35713/aic.v2.i6.79
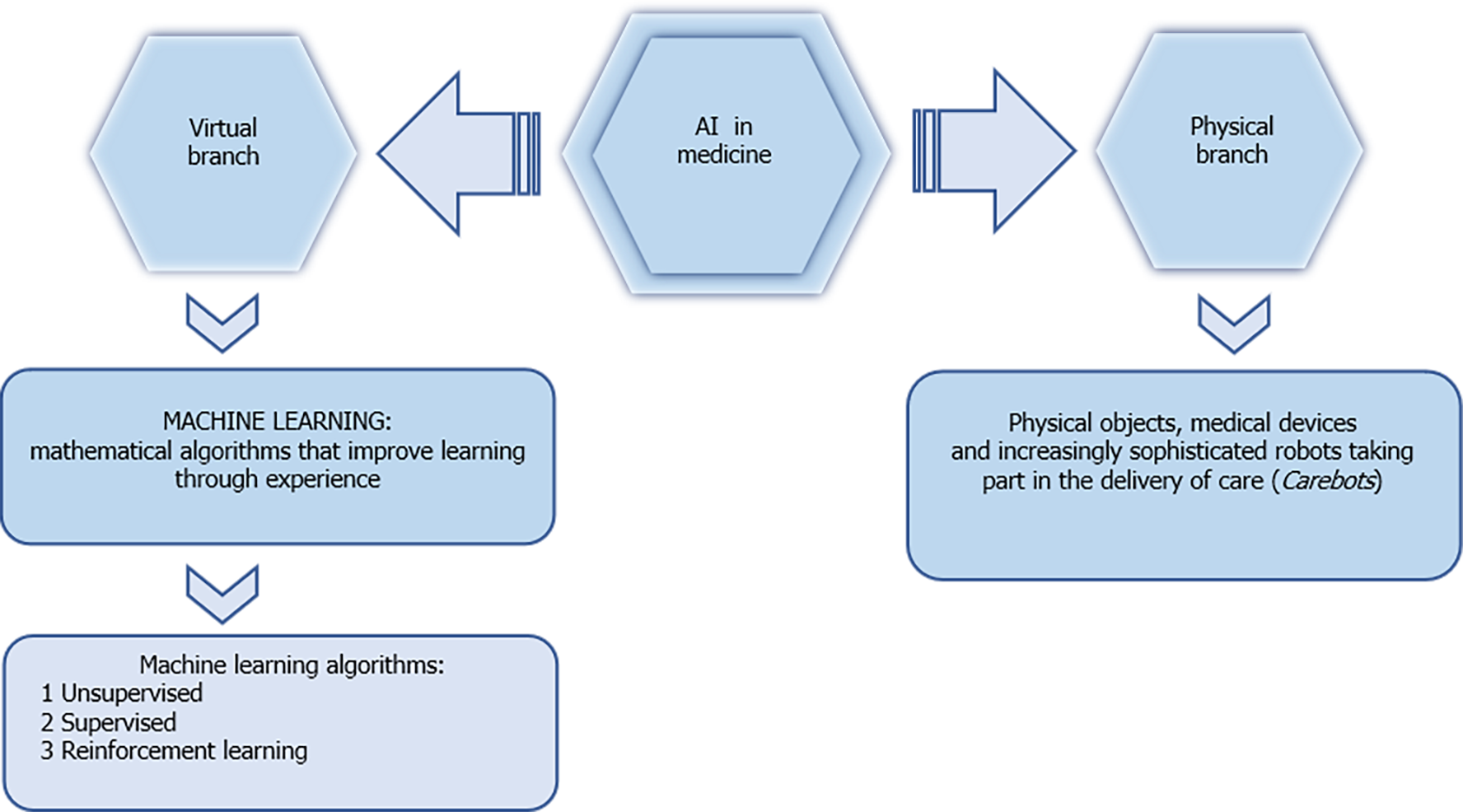
Colorectal cancer (CRC) is a growing disease around the world. It represents the third leading cause of cancer in males, the second in female patients, and fourth in the world for the cause of cancer death[1,2]. In 2015, 777987 new cases and 352589 deaths due to CRC were estimated in developed countries[3,4]. According to the Surveillance, Epidemiology, and End results program, the 5-year relative survival rate is between 63% and 67% for colon and rectal cancer, respectively[5]. Although important progress has been made in terms of understanding and treating CRC, morbidity and mortality rates based on recurrence and metastasis in therapy remain high[6-8]. The estimate of cases diagnosed at an advanced stage in asymptomatic patients is about 60%-70%[9-11]. High incidence, high mortality, and often poor prognosis make this disease not only a major health problem but also a social and economic one. For some years, early diagnosis and treatment of CRC patients have been the main clinical commitment. The research on the application of artificial intelligence (AI) in the treatment of CRC is still in its initial phase; however, with its continuous development and its major applications in the field of medicine, it is now taking off, also in the management of malignant colorectal disease. AI refers to a type of intelligence exhibited by machines that are able to perceive their environment and act autonomously in achieving their goals[12]. AI technology has been used extensively in medicine, business, and relationship life. In health science, AI is used especially for the diagnosis, treatment, and prognosis of diseases. In medicine, AI is divided into two sectors: Virtual and physical. The former includes imaging techniques, clinical assistant diagnosis, treatment, research, and drug development. The latter includes surgical and nursing robotic automation[13] (Figure 1). The continuous expansion and application of AI in the medical field are increasing its applicative prospects for the diagnosis and treatment of tumors. Recent studies have shown that AI can play an important role in the diagnosis and treatment of CRC patients, and it can not only improve the efficiency of screening, but can also improve the 5-year survival rate of CRC patients after treatment. This mini-review covers a period of 20 years with particular attention to the last decade, and intends to open a window on the attempts being made on the application of AI in the scientific and clinical research of the CRC by summarizing the most evident results. Our aim is not to draw definitive conclusions but to stimulate the interest of researchers in the application of these new technologies, which seem to be able to offer valuable help in the near future.
AI is an operative modality that can improve the collection of medical data for epidemiology purposes. Its predictive value is one of the most practical applications of this technology[14], already verified in areas such as public health and safety; on the other hand, it has not shown the same results when applied to the management of malignant diseases such as cancer. In 2016, CRC was responsible for approximately 8.2% of all cancer deaths in the United States, ranking as the third leading cause of cancer deaths regardless of gender[15]. In fact, we wonder how AI can positively influence the epidemiology of CRC. The first steps in epidemiology concern general measures such as the interpretation of research and data. There are often some difficulties regarding access to data and their categorization, and these methods are still under development[16-19]. Regarding CRC, an aspect of AI called GeoAI is a tool that can potentially help health care, and it is a system that collects information and data from a specific geographic area (food, type of soil, health system available, etc.) allowing them to be retrieved in a more specific and detailed form on the basis of what is desired[20]. Hungary and South Korea are the countries with the highest incidence of CRC[21], and CRC, like other gastrointestinal cancers, involves a complex and multifactorial process that is also influenced by geographic location, genetic predisposition, nutrition, lifestyles, and specific habits[22]. Due to this multifactoriality, systems such as GeoAI could have a great utility in defining more precisely the epidemiology of CRC by providing increasingly specific data sets in order to improve knowledge even at a local level. The hope is that specific areas with a high incidence of CRC will benefit from this type of information collection. Another application of AI is digital epidemiology which deals with the collection of information such as the collection of epidemiological data from social media and digital devices which are then quantified by AI[23]. Digital epidemiology has the great advantage of collecting huge amounts of data that have not been previously planned, but are instead voluntarily provided by people online[24]. This provides a huge pool of information that was not previously available to physicians and can aid in the early diagnosis of disease and health surveillance of the general public[25]. The disadvantage is the risk of violation of the patient's privacy which can bring out issues on information security and confidentiality[19,26]. Although the collection of information sets is considered tedious and not very useful, an advantageous tool for collecting and ordering them in order to produce something relevant is the data mining[27] that uses AI to collect much information, related or unrelated, trying to create a useful order to propose models and find facts[28]. This process involves the creation of databases, with selection and integration of data, storage and extraction of the most relevant ones, and proposal of models with subsequent evaluation and knowledge deriving from this information[28]. Therefore, medical data on CRC can come from different sources (social web, tertiary or research centers, etc.), but what is considered useful is the knowledge that they can bring and the use that can result from it. For CRC, data mining can represent a technological tool capable of implementing and promoting the discovery of relationships, new associations and other factors never even considered before which nevertheless may play a role in this multifactorial disease.
For many researchers, AI is seen as an approach that will help to better understand diseases and facilitate their management, and in this discourse it could not be excluded that slowly implemented ways can better diagnose cancer and treat patients with greater accuracy and precision[29]. Deep learning has attempted to examine how medical imaging can be improved and how to find cancer using imaging, and ranges from tools that enable a greater ability to scan and interpret images faster, high workflow, and better definition or improve image quality and its extraction with 3D technology[30-33]. Diagnosis is the integration of multi-source data analysis and clinical experience. Cancer manifests a wide variety of symptoms, rapid progression, drug susceptibility, and individual reactions, and for these reasons it is difficult to make an accurate diagnosis. AI has been shown to help clinicians in the qualitative diagnosis and staging of CRC[34].
Endoscopy is used to directly observe lesions in the intestinal wall, and endoscopists through images can assess whether the lesions are related to CRC. Lefere et al[35] in 2006 introduced the concept of virtual colonoscopy. This innovative examination is based on computed tomography colonography[36], in which the images are processed into three-dimensional cavity images. The images thus produce simulated optical colonoscopy with the aim of detecting CRCs and their adenomatoid polypoid precursors, or other neoplastic lesions. The advent of AI has made colonoscopy a convenient and accurate examination for CRC screening. In 2016, Fernandez-Esparrach et al[37] designed a method that automatically detected colon polyps. Their work achieved a sensitivity of 70.4% and specificity of 72.4%, and consisted of inserting 31 types of information about polyps into a computerized learning system. In 2012, Takemura et al[38] used narrow band imaging (NBI) and support vector machine (SVM) technology to distinguish neoplastic polyps from non-neoplastic polyps, resulting in a detection accuracy of 97.8%. Urban et al[39] designed and trained a convolutional neural network (CNN) system to improve adenoma recognition rate for colonoscopy. They collected images from over 2000 colonoscopy results for machine learning. Their assistant system achieved an accuracy of 96.4%. Mori et al[40] mixed NBI with staining image technology to recognize images of small malignant polyps being screened in real time. The final pathological forecast that they obtained was 98.1%. Akbari et al[41] used polyp segmentation during colonoscopy to recognize tumors using a CNN. During the testing phase, they conducted effective post-processing of a probability graph extrapolated from their CNN, reaching a specificity of 74.8%, sensitivity of 99.3%, and accuracy of 97.7%. Renner[42] have structured a computer-assisted optical biopsy system. They uploaded 602 images to the deep learning system for each colorectal tract examined endoscopically. By processing the information contained in the images, they were able to distinguish the neoplastic polyps with a diagnostic accuracy and sensitivity of 78.0% and 92.3%, respectively. Other authors[43], according to available evidence, conclude that the incorporation of AI as an aid for detection of colorectal neoplasia results in a significant increase in the detection of colorectal neoplasia, and such effect is independent from main adenoma characteristics. EndoBRAIN is an AI-assisted endoscopic diagnosis system that analyzes cell nuclei, crypt structures, and microvessels, with the aim of identifying colonic neoplasms. In 2020, Kudo et al[44] performed a retrospective comparison between the diagnostic capabilities of the EndoBRAIN system and those of 30 endoscopists. During the analysis, EndoBRAIN showed a sensitivity of 96.9%, specificity of 94.3%, and accuracy of 96.0% in distinguishing neoplastic from non-neoplastic lesions. The endoscopists' values were lower. Blanes-Vidal et al[45] extended the use of AI for capsule endoscopy through the use of a CNN for the detection and localization of colon polyps. The results of their algorithm were excellent, reaching an accuracy of 96.4%, while the sensitivity and specificity were 97.1% and 93.3%, respectively. During colonoscopy, the mucosa of malignant colon tumors is characterized by irregular and discontinuous crypt structures, and help in diagnosis can be provided by computer-assisted diagnosis (CAD). In 2015, the Infocus-Breakpoint was designed, which is a method that can directly detect the length and area of a neoplasm by transforming it into a 2D colonoscopic image, with great precision[46]. CAD was used by Ştefănescu[47] for processing images from confocal laser endomicroscopy and training the model using a two-layered feed forward neural network for diagnosing malignant samples based on seven parameters tested. The diagnostic error obtained was 15.5%. NBI magnification (M-NBI) can be employed for detailed observations of microvascular structures. In this regard, Tamai et al[48] used M-NBI-based CAD to classify and list mucosal lesions in the colon, including hyperplastic polyps, adenoma/adenocarcinoma lesions, and deep submucosal lesions, with an accuracy of 83.9%, 82.6%, 53.1%, 95.6%, and 82.8%, respectively. The development of CRC is a process consisting of many steps, and the transformation from adenoma to carcinoma[49] can take a very long time. Therefore, the importance of early screening and detection of lesions to reduce the incidence of this disease is intuitive[50]. In this regard, Ito et al[51] developed an AI system applied to endoscopy for diagnosis, and this system was based on a CNN using machine learning images. The authors analyzed protruding, flat, and sunken lesions and found improvement for colon cancer detection. The sensitivity, specificity, and accuracy found for cT1b were 67.5%, 89.0%, and 81.2%, respectively. Subsequently, several CAD systems were developed to screen patients at risk for CRC prior to colonoscopy. Some authors have developed AI systems with the aim of analyzing patient information comprehensively to predict the onset of CRC. The variables selected were gender, age, and blood test data. In this case, the objective in addition to the scientific purpose represented an encouragement for patients with positive results in order to induce them to accept periodic checks[52]. In 2018, Xu et al[53] assembled a team that designed an early screening method for CRC based on plasma copy number variation. They sequenced entire genomes and then trained an SVM to make the diagnosis. The results obtained by this method were an 88.9% specificity and a sensitivity of 91.7%. Recently, Graham et al[54] proposed MILD-Net, a CNN composed of a completely convolutive network, and this system has reintroduced the original images at multiple points within their network in order to reduce diagnostic uncertainty. Other authors such as Wan et al[55] designed an AI program with the intent of improving the sensitivity of the extraction of plasma cell-free DNA for CRC patients. For a weighted early (stage I/II) CRC cohort, they achieved an average sensitivity of 85%. Wang et al[56] has designed several artificial neural networks (ANN) models that are biologically inspired computer programs designed to simulate the way in which the human brain processes information. Using a vector quantization neural network, they structured four models for qualitative diagnosis, M0/M1 discrimination, carcinoembryonic antigen testing, and clinical staging. Shahbazy et al[57] have included some classification factors in his algorithm, concerning a case-control study, demonstrating greater accuracy in the early diagnosis of CRC. The 5-year disease-free survival rate was 84%. Tumor budding is considered a sign of cancer cell activity and the first step of tumor metastasis. In accordance with this concept, Liu[58] in 2021 established an automatic diagnostic platform for rectal cancer budding pathology by training a faster region-based CNN (F-R-CNN) on the pathological images of rectal cancer budding. He analyzed postoperative pathological section images of 236 patients with rectal cancer. The conclusions were that F-R-CNN deep neural network platform for the pathological diagnosis of rectal cancer tumor budding can help pathologists make more efficient and accurate pathological diagnoses. Gupta et al[59] proposed a study on over 4000 CRC patients, using machine learning algorithms to predict the stage of the tumor. They postulated that tumor budding may be an additional prognostic factor to the TNM staging system. To overcome the problem of the poor reliability of tumor budding, Weis et al[60] introduced a different automatic image processing tool to quantify it in immunohistochemistry sections. Detections of tumor buds in CRC patient samples were reliable. To increase the results of AI against CRC, histopathology and genetics may not be enough, in fact, as shown by a study by Borkowski et al[61] who compared different AI platforms to detect adenocarcinoma in the veteran population. They found significant difficulty, on the part of machine learning tools, in differentiating adenocarcinoma with KRAS mutation from those without KRAS mutation. These difficulties may suggest the need for a more unified approach. Environmental causes should not be underestimated in the formation of CRC cancer. However, the study of tumor suppressor genes and oncogenes such as APC, KRAS, and MTHFR occupy a no less important role[62]. This has led to a dramatic increase in the number of potential biomarkers and indicators that can be linked to cancer growth. Some success has been achieved through AI training to classify tumors based on histopathology alone. Some authors[63] have trained an algorithm to classify gastric and epithelial tumors into adenocarcinoma, adenoma, or non-neoplastic lesions. They compared a type of CNN that uses smaller tile sizes to RNN (recurrent neural network) to observe and then classify the images. The RNN was more accurate and no statistical difference was demonstrated. Ciompi et al[64] postulated that CNNs need better image quality and this could help AI approach. Late diagnosis of CRC can lead to other problems and we are nowhere near the solution, but AI could help in other ways for CRC diagnosis at a later stage. A study by Dimitriou et al[65] sought to increase the accuracy in prognosis for stage II cancers, ultimately arguing that more attention should be paid to traits such as textures, spatial relationships, and morphology, all of which can be better managed by machine learning. Another tool that could be useful in diagnosing CRC is represented by fuzzy systems[66], and these allow any number between 0 and 1 to exist. This scope can be applied to any form of pathology and can help identify aberrations or predict the likelihood of cancer based on predetermined parameters. In the case of colorectal polyps, fuzzy parameters can be easily applied to match various characteristics as a stock of polyps in pedunculate or mucous lesions, if the indentations on hyperplastic polyps will become advanced lesions and specific sizes or forms, and to categorize low-risk injuries or if an adenomatous polyp is at risk of canceration.
AI can also be important in genetic testing for CRC. Hu et al[67], based on gene expression, compared the accuracies of three different neural networks for cancer classification. On a sample of 53 patients, they found that the most accurate classification was obtained with the S-Kohonen neural network. In 2017, Xu et al[68] structured an SVM system to identify differentially expressed genes with the purpose of distinguishing patients with high risk and predicting prognoses. Fifteen genetic markers were identified as predictors of recurrence risk and prognosis for colon cancer patients. Zhang et al[69] developed a counter-propagation ANN for the detection of the BRAF gene mutation in CRC using near-infrared testing. Their model achieved a diagnostic sensitivity of 100%, specificity of 87.5%, and accuracy of 93.8%, and it can distinguish the BRAF V600E mutation from the wild type.
Methylated DNA has been widely used in AI diagnosis as a biomarker for early CRC. Coppede et al[70] in 2015 structured an ANN to explore the association between CRC-related genes and environmental factors. They concluded that ANNs revealed the complexity of the interconnections among factors linked to DNA methylation in CRC, and also observed an intricate network of interconnections between dietary and lifestyle factors and the methylation profiles of the studied genes. Kel et al[71] developed an analytical method to diagnose early CRC by extracting human methylated cytosine and guanine separated by a phosphate (CpG) from blood and feces. This study involved 300 CRC patients and identified six potential epigenetic biomarkers of DNA methylation that may lead to rapid tumor development.
In 2019 Ferrari et al[72] reported that AI, based on the analysis of the texture of MR images, would be able to settle the complete therapeutic response of rectal cancer previously subjected to neoadjuvant chemotherapy. Their results have been encouraging. Indeed, the proposed AI model allows the distinction between complete response to therapy and non-response in neoadjuvant treatment of rectal cancer. The role of AI in drug metabolism in the treatment of CRC is not to be neglected, and it can allow a better understanding of the transformation and metabolism that drugs induce towards cancer progression. Tools that assist AI provide reliable information on the metabolism of these drugs in the treatment of CRC, leading to a better understanding of their biological behavior and specific metabolic pathways[73]. The predictive power of AI in CRC through the use of the ANN algorithm is increasingly appreciated. Indeed, the ANN uses non-linear models with particular flexibility regarding medical research and clinical practice[74,75]. An advantage could be to optimize the process through flexible models with good value for money, and for large data collections. The ANN has proven to be an accurate and reliable tool for clinical decision making. Lastly, academic dissemination of knowledge is facilitated by these models[74-78]. According to a systematic review of 27 studies that used ANNs as diagnostic or prognostic tools, 21 of these showed health care benefits, while the others showed similar results to models already in use[74]. In this regard, other authors reported that the ANN applied to the prediction of distant metastasis of CRC showed a better outcome[79]. Traditionally, the treatment of CRC is multimodal, integrating surgery, chemotherapy, radiotherapy, and immunotherapy, and aims to offer together a complete and more effective cure. AI can be an extra help for patients to choose the treatment methods that are appropriate for them and improve the healing effects of the protocols in use by designing more individualized and precise therapies.
Cancer research is moving towards the personalization of cancer treatments. Healthcare is rapidly moving toward precision or personalized medicine. Machine learning models have the potential to contribute to individual-based cancer care and transform the future of medicine[80]. The Watson for Oncology system was developed at Memorial Sloan Kettering Cancer Center. This AI-based system, by automatically extracting medical information from medical records and translating it into a practical language for learning, improves personalized and precision medical treatment for cancer therapy. This tool has been evaluated by various authors. Kim in 2019[81] analyzed the concordance rate between different chemotherapy regimens in the treatment of CRC, and the data were compared between those determined by a multidisciplinary team and those suggested by the recommendations of the Watson for Oncology: In 61 CRC samples, the rate of initial agreement was 46.4%, but after inclusion of other recommendations, it increased to 88.4%. It would appear that this system can be improved through continuous adjustments. Miyano[82] has also used the Watson for Oncology system for the genetic sequencing of cancer patients, and the results obtained in a very rapid time have produced a reduction in waiting times, which is very useful when the topic under discussion is cancer. Akturk and Erci[83] experimented with this model applied to human care, finding more individualized and caring nursing services with patient satisfaction. With regard to these pathologies, medicine is becoming more and more personalized and also for the drugs used. In this sense, Keshava et al[84] proposed a method that can identify subpopulations that have different reactions towards inhibitors of the same target and can help to understand the mechanisms of resistance. With a system in continuous information enrichment, new subpopulations of cancer could be identified, and old and new genetic biomarkers were analyzed in order to find more effective combinations of drugs. Still with regard to the application of AI on targeted drugs, Ding et al[85] created a system for screening molecular markers at the system biology level by integrating transcriptomics and proteomics data. The identified markers were integrated to develop targeted drugs useful for the clinical treatment of CRC. Nowak-Sliwinska et al[86] proposed a study using existing anticancer drugs to treat new indications. They combined specific phenotypic studies with mechanistic studies to create AI models capable of predicting disease-drug pairs. Finally, clinical management also cannot be neglected. Horta et al[87] in 2018 collected information from CRC surgical patients, and then instructed a model to support decisions regarding selection of patients who should be offered co-management services.
With regard to new technologies such as AI, there is often a doubtful attitude, and the health staff bases their work and commitment on certain and verified information. However, in the era of big data, it will be necessary to address these issues and certainly deepen them in order to understand how reliable they are and the help that these methods can give us. The next step should be the creation of medical ethics guidelines, in order to regulate the scope of the use of these new technologies. It is necessary to understand the data in order to draw firm conclusions. The limit of all this technology is the insufficiency of aggregate and understandable data that allow us to draw advantageous conclusions. Taking the studies on the subject of CT imaging and AI as an example, the input data can be manifold, and this can represent a problem when definitive answers must be given for the diagnosis and treatment of the general population. The data optimization that the new technologies offer us requires and will require an investment of more and more time and money, but this will allow us to build systems that will allow better data collection and that will allow better and more accurate decision-making processes. The more institutions that start accumulating data, the greater their quantity and quality. The creation of public databases for information such as symptomatology, different imaging modalities, or geographic distribution can be a great strength that could benefit researchers by having access to more and more information. Free access to this data represents another obstacle that should not be underestimated, and underdeveloped and poor countries may not be able to have access to this technology. The further on we will go with the experimentation and application of AI, the more costs will decrease and the greater will be the benefits that can be felt by everyone and not only by those in certain geographical areas. It is critical for the global health community that these countries have access to technology to better treat and address diseases in their area and improve the quality of life in their local communities.
All new technologies can represent the beginning of a new era and their applications in daily use take time to be slowly incorporated. AI is certainly a promise of a new scientific season, but it remains at an early stage for its true application. Several researches on their use are slowly moving in a good direction. To date, it is clear that the methods of information gathering, diagnosis, and treatment of CRC will greatly improve through the use of deep learning tools. While the methods of obtaining medical information may be controversial, we can say with certainty that early detection of CRC will gain a great deal when appropriate methods for data collection are found. Ultimately, AI shows great promise in clinical and therapeutic management for CRC, and this could indicate better and more personalized treatments for patients with this disease.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: Fellow of American College of surgeons, 03355092.
Specialty type: Surgery
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Gao W, Naik N, Tanabe S S-Editor: Liu M L-Editor: Wang TQ P-Editor: Liu M
| 1. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21370] [Article Influence: 2137.0] [Reference Citation Analysis (3)] |
| 2. | Maida M, Morreale G, Sinagra E, Ianiro G, Margherita V, Cirrone Cipolla A, Camilleri S. Quality measures improving endoscopic screening of colorectal cancer: a review of the literature. Expert Rev Anticancer Ther. 2019;19:223-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (1)] |
| 3. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11128] [Cited by in RCA: 11836] [Article Influence: 845.4] [Reference Citation Analysis (4)] |
| 4. | He X, Li S, Yu B, Kuang G, Wu Y, Zhang M, He Y, Ou C, Cao P. Up-regulation of LINC00467 promotes the tumourigenesis in colorectal cancer. J Cancer. 2019;10:6405-6413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 5. | American Cancer Society. Survival Rates for Colorectal Cancer. [cited 15 March 2021]. Available from: https://www.cancer.org/cancer/colon-rectal-cancer/detection-diagnosis-staging/survival-rates.html. |
| 6. | Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2526] [Cited by in RCA: 2912] [Article Influence: 364.0] [Reference Citation Analysis (3)] |
| 7. | Wang Y, Nie H, He X, Liao Z, Zhou Y, Zhou J, Ou C. The emerging role of super enhancer-derived noncoding RNAs in human cancer. Theranostics. 2020;10:11049-11062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (1)] |
| 8. | Ou C, Sun Z, Li S, Li G, Li X, Ma J. Dual roles of yes-associated protein (YAP) in colorectal cancer. Oncotarget. 2017;8:75727-75741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 9. | Mandel JS, Bond JH, Church TR, Snover DC, Bradley GM, Schuman LM, Ederer F. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study. N Engl J Med. 1993;328:1365-1371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2174] [Article Influence: 67.9] [Reference Citation Analysis (1)] |
| 10. | Maida M, Macaluso FS, Ianiro G, Mangiola F, Sinagra E, Hold G, Maida C, Cammarota G, Gasbarrini A, Scarpulla G. Screening of colorectal cancer: present and future. Expert Rev Anticancer Ther. 2017;17:1131-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 11. | Sinagra E, Badalamenti M, Maida M, Spadaccini M, Maselli R, Rossi F, Conoscenti G, Raimondo D, Pallio S, Repici A, Anderloni A. Use of artificial intelligence in improving adenoma detection rate during colonoscopy: Might both endoscopists and pathologists be further helped. World J Gastroenterol. 2020;26:5911-5918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 12. | Francolini G, Desideri I, Stocchi G, Salvestrini V, Ciccone LP, Garlatti P, Loi M, Livi L. Artificial Intelligence in radiotherapy: state of the art and future directions. Med Oncol. 2020;37:50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 13. | Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism. 2017;69S:S36-S40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 832] [Article Influence: 104.0] [Reference Citation Analysis (0)] |
| 14. | Chen JH, Asch SM. Machine Learning and Prediction in Medicine - Beyond the Peak of Inflated Expectations. N Engl J Med. 2017;376:2507-2509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 547] [Cited by in RCA: 602] [Article Influence: 75.3] [Reference Citation Analysis (4)] |
| 15. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12135] [Cited by in RCA: 12991] [Article Influence: 1443.4] [Reference Citation Analysis (2)] |
| 16. | Thiébaut R, Cossin S; Section Editors for the IMIA Yearbook Section on Public Health and Epidemiology Informatics. Artificial Intelligence for Surveillance in Public Health. Yearb Med Inform. 2019;28:232-234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 17. | Thiébaut R, Thiessard F; Section Editors for the IMIA Yearbook Section on Public Health and Epidemiology Informatics. Artificial Intelligence in Public Health and Epidemiology. Yearb Med Inform. 2018;27:207-210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 18. | Rodríguez-González A, Zanin M, Menasalvas-Ruiz E. Public Health and Epidemiology Informatics: Can Artificial Intelligence Help Future Global Challenges? Yearb Med Inform. 2019;28:224-231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Denecke K. An ethical assessment model for digital disease detection technologies. Life Sci Soc Policy. 2017;13:16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 20. | Janowicz K, Gao S, McKenzie G, Hu Y, Bhaduri B. GeoAI: Spatially Explicit Artificial Intelligence Techniques for Geographic Knowledge Discovery and Beyond. Int J Geogr Inf Sci. 2020;625-636. [DOI] [Full Text] |
| 21. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55838] [Article Influence: 7976.9] [Reference Citation Analysis (132)] |
| 22. | Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1014] [Cited by in RCA: 1122] [Article Influence: 112.2] [Reference Citation Analysis (0)] |
| 23. | Velasco E. Disease detection, epidemiology and outbreak response: the digital future of public health practice. Life Sci Soc Policy. 2018;14:7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 24. | Tarkoma S, Alghnam S, Howell MD. Fighting pandemics with digital epidemiology. EClinicalMedicine. 2020;26:100512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Eckmanns T, Füller H, Roberts SL. Digital epidemiology and global health security; an interdisciplinary conversation. Life Sci Soc Policy. 2019;15:2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 26. | Kostkova P. Disease surveillance data sharing for public health: the next ethical frontiers. Life Sci Soc Policy. 2018;14:16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 27. | Hand D, Mannila H, Smyth P. Principles of Data Mining. Cambridge (MA): MIT Press, 2001: 546. |
| 28. | Han J, Kamber M, Pei J. Data mining: Concepts and techniques. In: The Morgan Kaufmann Series in Data Management Systems. 3rd ed. Waltham, 2012. |
| 29. | Huang S, Yang J, Fong S, Zhao Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020;471:61-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 296] [Article Influence: 49.3] [Reference Citation Analysis (1)] |
| 30. | Liu J, Pan Y, Li M, Chen Z, Tang L, Lu C, Wang J. Applications of deep learning to MRI images: A survey, in Big Data Mining and Analytics. Big Data Min Anal. 2018;1:1-18. [RCA] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 141] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 31. | Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2376] [Cited by in RCA: 2837] [Article Influence: 472.8] [Reference Citation Analysis (0)] |
| 32. | Thompson RF, Valdes G, Fuller CD, Carpenter CM, Morin O, Aneja S, Lindsay WD, Aerts HJWL, Agrimson B, Deville C Jr, Rosenthal SA, Yu JB, Thomas CR Jr. Artificial intelligence in radiation oncology: A specialty-wide disruptive transformation? Radiother Oncol. 2018;129:421-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 150] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 33. | Li M, Shen S, Gao W, Hsu W, Cong J. Computed Tomography Image Enhancement Using 3D Convolutional Neural Network. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Switzerland: Springer Nature, 2018. [DOI] [Full Text] |
| 34. | Gupta N, Kupfer SS, Davis AM. Colorectal Cancer Screening. JAMA. 2019;321:2022-2023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 35. | Lefere P, Gryspeerdt S, Schotte K. Virtual colonoscopy--an overview. Onkologie. 2006;29:281-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 36. | Halligan S, Altman DG, Taylor SA, Mallett S, Deeks JJ, Bartram CI, Atkin W. CT colonography in the detection of colorectal polyps and cancer: systematic review, meta-analysis, and proposed minimum data set for study level reporting. Radiology. 2005;237:893-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 242] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 37. | Fernández-Esparrach G, Bernal J, López-Cerón M, Córdova H, Sánchez-Montes C, Rodríguez de Miguel C, Sánchez FJ. Exploring the clinical potential of an automatic colonic polyp detection method based on the creation of energy maps. Endoscopy. 2016;48:837-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 38. | Takemura Y, Yoshida S, Tanaka S, Kawase R, Onji K, Oka S, Tamaki T, Raytchev B, Kaneda K, Yoshihara M, Chayama K. Computer-aided system for predicting the histology of colorectal tumors by using narrow-band imaging magnifying colonoscopy (with video). Gastrointest Endosc. 2012;75:179-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 39. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology 2018; 155: 1069-1078. e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 433] [Article Influence: 61.9] [Reference Citation Analysis (1)] |
| 40. | Mori Y, Kudo SE, Misawa M, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y, Maeda Y, Takeda K, Nakamura H, Ichimasa K, Kudo T, Hayashi T, Wakamura K, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 355] [Article Influence: 50.7] [Reference Citation Analysis (1)] |
| 41. | Akbari M, Mohrekesh M, Nasr-Esfahani E, Soroushmehr SMR, Karimi N, Samavi S, Najarian K. Polyp Segmentation in Colonoscopy Images Using Fully Convolutional Network. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:69-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 42. | Renner J, Phlipsen H, Haller B, Navarro-Avila F, Saint-Hill-Febles Y, Mateus D, Ponchon T, Poszler A, Abdelhafez M, Schmid RM, von Delius S, Klare P. Optical classification of neoplastic colorectal polyps - a computer-assisted approach (the COACH study). Scand J Gastroenterol. 2018;53:1100-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Hassan C, Spadaccini M, Iannone A, Maselli R, Jovani M, Chandrasekar VT, Antonelli G, Yu H, Areia M, Dinis-Ribeiro M, Bhandari P, Sharma P, Rex DK, Rösch T, Wallace M, Repici A. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastrointest Endosc 2021; 93: 77-85. e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 308] [Article Influence: 77.0] [Reference Citation Analysis (1)] |
| 44. | Kudo SE, Misawa M, Mori Y, Hotta K, Ohtsuka K, Ikematsu H, Saito Y, Takeda K, Nakamura H, Ichimasa K, Ishigaki T, Toyoshima N, Kudo T, Hayashi T, Wakamura K, Baba T, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Artificial Intelligence-assisted System Improves Endoscopic Identification of Colorectal Neoplasms. Clin Gastroenterol Hepatol 2020; 18: 1874-1881. e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 45. | Blanes-Vidal V, Baatrup G, Nadimi ES. Addressing priority challenges in the detection and assessment of colorectal polyps from capsule endoscopy and colonoscopy in colorectal cancer screening using machine learning. Acta Oncol. 2019;58:S29-S36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 46. | Chadebecq F, Tilmant C, Bartoli A. How big is this neoplasia? Med Image Anal. 2015;19:58-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 47. | Ştefănescu D, Streba C, Cârţână ET, Săftoiu A, Gruionu G, Gruionu LG. Computer Aided Diagnosis for Confocal Laser Endomicroscopy in Advanced Colorectal Adenocarcinoma. PLoS One. 2016;11:e0154863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 48. | Tamai N, Saito Y, Sakamoto T, Nakajima T, Matsuda T, Sumiyama K, Tajiri H, Koyama R, Kido S. Effectiveness of computer-aided diagnosis of colorectal lesions using novel software for magnifying narrow-band imaging: a pilot study. Endosc Int Open. 2017;5:E690-E694. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 49. | Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490-1502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1965] [Cited by in RCA: 2294] [Article Influence: 208.5] [Reference Citation Analysis (1)] |
| 50. | Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967-976. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 245] [Cited by in RCA: 428] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 51. | Ito N, Kawahira H, Nakashima H, Uesato M, Miyauchi H, Matsubara H. Endoscopic Diagnostic Support System for cT1b Colorectal Cancer Using Deep Learning. Oncology. 2019;96:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 52. | Hornbrook MC, Goshen R, Choman E, O'Keeffe-Rosetti M, Kinar Y, Liles EG, Rust KC. Early Colorectal Cancer Detected by Machine Learning Model Using Gender, Age, and Complete Blood Count Data. Dig Dis Sci. 2017;62:2719-2727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 53. | Xu JF, Kang Q, Ma XY, Pan YM, Yang L, Jin P, Wang X, Li CG, Chen XC, Wu C, Jiao SZ, Sheng JQ. A Novel Method to Detect Early Colorectal Cancer Based on Chromosome Copy Number Variation in Plasma. Cell Physiol Biochem. 2018;45:1444-1454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 54. | Graham S, Chen H, Gamper J, Dou Q, Heng PA, Snead D, Tsang YW, Rajpoot N. MILD-Net: Minimal information loss dilated network for gland instance segmentation in colon histology images. Med Image Anal. 2019;52:199-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 129] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 55. | Wan N, Weinberg D, Liu TY, Niehaus K, Ariazi EA, Delubac D, Kannan A, White B, Bailey M, Bertin M, Boley N, Bowen D, Cregg J, Drake AM, Ennis R, Fransen S, Gafni E, Hansen L, Liu Y, Otte GL, Pecson J, Rice B, Sanderson GE, Sharma A, St John J, Tang C, Tzou A, Young L, Putcha G, Haque IS. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer. 2019;19:832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 118] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 56. | Wang Q, Wei J, Chen Z, Zhang T, Zhong J, Zhong B, Yang P, Li W, Cao J. Establishment of multiple diagnosis models for colorectal cancer with artificial neural networks. Oncol Lett. 2019;17:3314-3322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 57. | Shahbazy M, Vasighi M, Kompany-Zareh M, Ballabio D. Oblique rotation of factors: a novel pattern recognition strategy to classify fluorescence excitation-emission matrices of human blood plasma for early diagnosis of colorectal cancer. Mol Biosyst. 2016;12:1963-1975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 58. | Liu S, Zhang Y, Ju Y, Li Y, Kang X, Yang X, Niu T, Xing X, Lu Y. Establishment and Clinical Application of an Artificial Intelligence Diagnostic Platform for Identifying Rectal Cancer Tumor Budding. Front Oncol. 2021;11:626626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (1)] |
| 59. | Gupta P, Chiang SF, Sahoo PK, Mohapatra SK, You JF, Onthoni DD, Hung HY, Chiang JM, Huang Y, Tsai WS. Prediction of Colon Cancer Stages and Survival Period with Machine Learning Approach. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 60. | Weis CA, Kather JN, Melchers S, Al-Ahmdi H, Pollheimer MJ, Langner C, Gaiser T. Automatic evaluation of tumor budding in immunohistochemically stained colorectal carcinomas and correlation to clinical outcome. Diagn Pathol. 2018;13:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 61. | Borkowski AA, Wilson CP, Borkowski SA, Thomas LB, Deland LA, Grewe SJ, Mastorides SM. Comparing Artificial Intelligence Platforms for Histopathologic Cancer Diagnosis. Fed Pract. 2019;36:456-463. [PubMed] |
| 62. | Hilario M, Kalousis A. Approaches to dimensionality reduction in proteomic biomarker studies. Brief Bioinform. 2008;9:102-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 98] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 63. | Komeda Y, Handa H, Watanabe T, Nomura T, Kitahashi M, Sakurai T, Okamoto A, Minami T, Kono M, Arizumi T, Takenaka M, Hagiwara S, Matsui S, Nishida N, Kashida H, Kudo M. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology. 2017;93 Suppl 1:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 64. | Ciompi F, Geessink O, Bejnordi BE, De Souza GS, Baidoshvili A, Litjens G, van Ginnekan B, Nagtegaal I, Van Der Laak J. The importance of stain normalization in colorectal tissue classification with convolutional networks. IEEE. 2017;160-163. [DOI] [Full Text] |
| 65. | Dimitriou N, Arandjelović O, Harrison DJ, Caie PD. A principled machine learning framework improves accuracy of stage II colorectal cancer prognosis. NPJ Digit Med. 2018;1:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 66. | Ngan TT, Lan LTH, Tuan TM, Son LH, Tuan LM, Minh NH. Colorectal Cancer Diagnosis with Complex Fuzzy Inference System. In: Frontiers in Intelligent Computing: Theory and Applications. Advances in Intelligent Systems and Computing 2020. Satapathy S, Bhateja V, Nguyen B, Nguyen N, Le DN, editors. Springer, Singapore, 2020. [DOI] [Full Text] |
| 67. | Hu HP, Niu ZJ, Bai YP, Tan XH. Cancer classification based on gene expression using neural networks. Genet Mol Res. 2015;14:17605-17611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 68. | Xu G, Zhang M, Zhu H, Xu J. A 15-gene signature for prediction of colon cancer recurrence and prognosis based on SVM. Gene. 2017;604:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 69. | Zhang X, Yang Y, Wang Y, Fan Q. Detection of the BRAF V600E Mutation in Colorectal Cancer by NIR Spectroscopy in Conjunction with Counter Propagation Artificial Neural Network. Molecules. 2019;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 70. | Coppedè F, Grossi E, Lopomo A, Spisni R, Buscema M, Migliore L. Application of artificial neural networks to link genetic and environmental factors to DNA methylation in colorectal cancer. Epigenomics. 2015;7:175-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 71. | Kel A, Boyarskikh U, Stegmaier P, Leskov LS, Sokolov AV, Yevshin I, Mandrik N, Stelmashenko D, Koschmann J, Kel-Margoulis O, Krull M, Martínez-Cardús A, Moran S, Esteller M, Kolpakov F, Filipenko M, Wingender E. Walking pathways with positive feedback loops reveal DNA methylation biomarkers of colorectal cancer. BMC Bioinformatics. 2019;20:119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 72. | Ferrari R, Mancini-Terracciano C, Voena C, Rengo M, Zerunian M, Ciardiello A, Grasso S, Mare' V, Paramatti R, Russomando A, Santacesaria R, Satta A, Solfaroli Camillocci E, Faccini R, Laghi A. MR-based artificial intelligence model to assess response to therapy in locally advanced rectal cancer. Eur J Radiol. 2019;118:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 73. | Lisboa PJ, Taktak AF. The use of artificial neural networks in decision support in cancer: a systematic review. Neural Netw. 2006;19:408-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 178] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 74. | Kickingereder P, Isensee F, Tursunova I, Petersen J, Neuberger U, Bonekamp D, Brugnara G, Schell M, Kessler T, Foltyn M, Harting I, Sahm F, Prager M, Nowosielski M, Wick A, Nolden M, Radbruch A, Debus J, Schlemmer HP, Heiland S, Platten M, von Deimling A, van den Bent MJ, Gorlia T, Wick W, Bendszus M, Maier-Hein KH. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol. 2019;20:728-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 288] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 75. | Yin M, Ma J, Xu J, Li L, Chen G, Sun Z, Liu Y, He S, Ye J, Mo W. Use of artificial neural networks to identify the predictive factors of extracorporeal shock wave therapy treating patients with chronic plantar fasciitis. Sci Rep. 2019;9:4207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 76. | Chen J, Remulla D, Nguyen JH, Aastha D, Liu Y, Dasgupta P, Hung AJ. Current status of artificial intelligence applications in urology and their potential to influence clinical practice. BJU Int. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 77. | Somashekhar SP, Sepúlveda MJ, Puglielli S, Norden AD, Shortliffe EH, Rohit Kumar C, Rauthan A, Arun Kumar N, Patil P, Rhee K, Ramya Y. Watson for Oncology and breast cancer treatment recommendations: agreement with an expert multidisciplinary tumor board. Ann Oncol. 2018;29:418-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 171] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 78. | Mayo RM, Summey JF, Williams JE, Spence RA, Kim S, Jagsi R. Qualitative Study of Oncologists' Views on the CancerLinQ Rapid Learning System. J Oncol Pract. 2017;13:e176-e184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 79. | Schöllhorn WI. Applications of artificial neural nets in clinical biomechanics. Clin Biomech (Bristol, Avon). 2004;19:876-898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 82] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 80. | Mitsala A, Tsalikidis C, Pitiakoudis M, Simopoulos C, Tsaroucha AK. Artificial Intelligence in Colorectal Cancer Screening, Diagnosis and Treatment. A New Era. Curr Oncol. 2021;28:1581-1607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 131] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 81. | Kim EJ, Woo HS, Cho JH, Sym SJ, Baek JH, Lee WS, Kwon KA, Kim KO, Chung JW, Park DK, Kim YJ. Early experience with Watson for oncology in Korean patients with colorectal cancer. PLoS One. 2019;14:e0213640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 82. | Miyano S. [Artificial Intelligence for Cancer Genomic Medicine: Understanding Cancer is Beyond Human Ability]. Brain Nerve. 2019;71:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 83. | Aktürk Ü, Erci B. The Effect of Watson's Human Caring Model on Meaning of Life and Symptom Management in Cancer Patients Undergoing Chemotherapy. Res Theory Nurs Pract. 2018;32:255-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 84. | Keshava N, Toh TS, Yuan H, Yang B, Menden MP, Wang D. Defining subpopulations of differential drug response to reveal novel target populations. NPJ Syst Biol Appl. 2019;5:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 85. | Ding D, Han S, Zhang H, He Y, Li Y. Predictive biomarkers of colorectal cancer. Comput Biol Chem. 2019;83:107106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 86. | Nowak-Sliwinska P, Scapozza L, Ruiz i Altaba A. Drug repurposing in oncology: Compounds, pathways, phenotypes and computational approaches for colorectal cancer. Biochim Biophys Acta Rev Cancer. 2019;1871:434-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 112] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 87. | Horta AB, Salgado C, Fernandes M, Vieira S, Sousa JM, Papoila AL, Xavier M. Clinical decision support tool for Co-management signalling. Int J Med Inform. 2018;113:56-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |