Published online Nov 28, 2020. doi: 10.35713/aic.v1.i4.51
Peer-review started: September 26, 2020
First decision: October 22, 2020
Revised: November 5, 2020
Accepted: November 21, 2020
Article in press: November 21, 2020
Published online: November 28, 2020
Processing time: 64 Days and 19.3 Hours
Understanding of the cellular signaling pathways involved in cancer disease is of great importance. These complex biological mechanisms can be thoroughly revealed by their structure, dynamics, and control methods. Artificial intelligence offers rule-based models that favor the research of human signaling processes. In this paper, we give an overview of the advantages of the formalism of symbolic models in medical biology and cell biology of the uveal melanoma. A language is described that allows us: (1) To define the system states and elements with their alterations; (2) To model the dynamics of the cellular system; and (3) To perform inference-based analysis with the logical tools of the language.
Core Tip: Artificial intelligence offers rule-based models that favor the understanding of cell biology (signaling pathways) involved in the uveal melanoma.
- Citation: Santos-Buitrago B, Santos-García G, Hernández-Galilea E. Artificial intelligence for modeling uveal melanoma. Artif Intell Cancer 2020; 1(4): 51-65
- URL: https://www.wjgnet.com/2644-3228/full/v1/i4/51.htm
- DOI: https://dx.doi.org/10.35713/aic.v1.i4.51
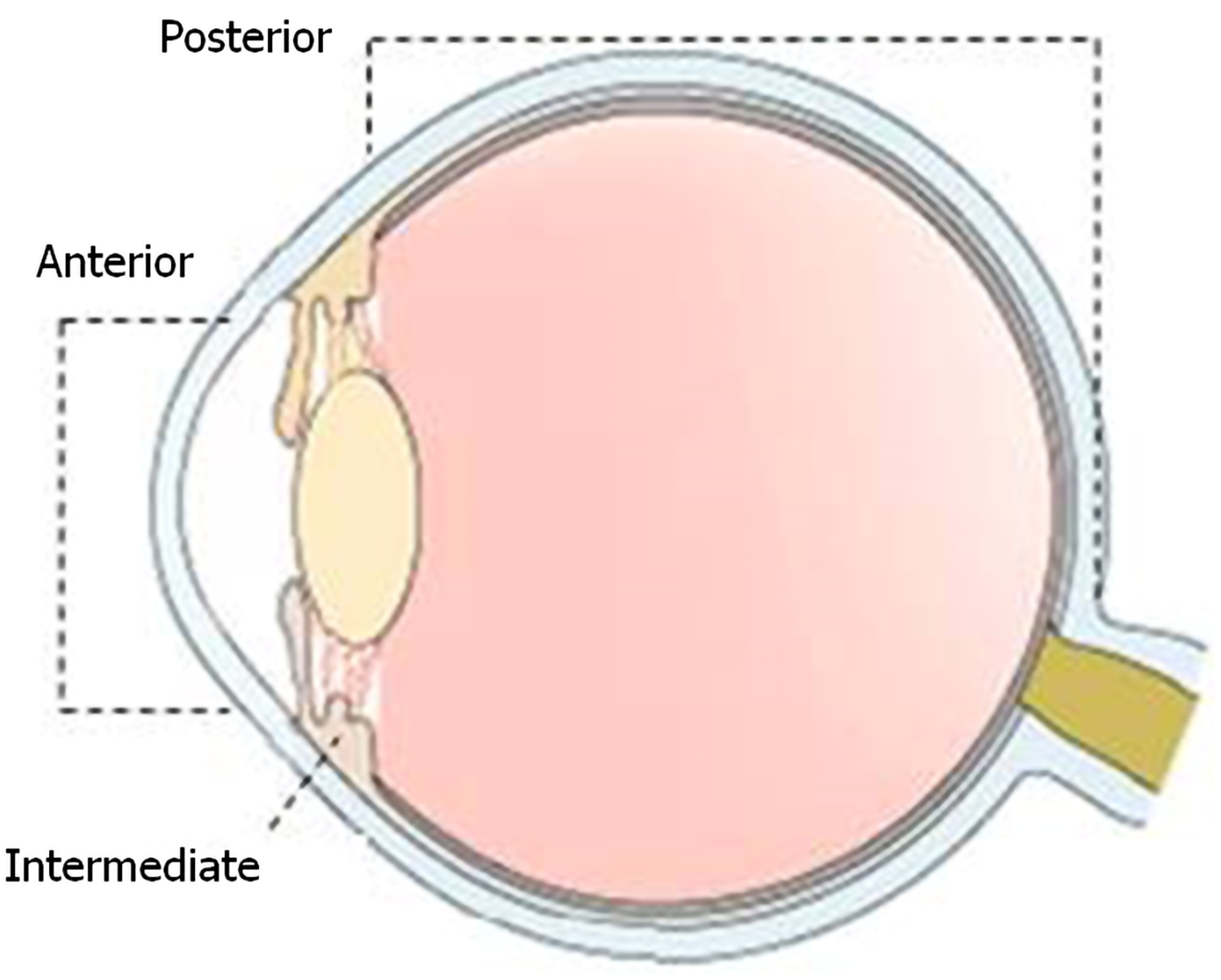
Regarding the eye anatomy, the uvea is the middle layer of the eyeball, also known as the vascular tunic, uveal layer, uveal coat, or uveal tract. It consists of three parts: the iris, the ciliary body, and the choroid (Figure 1). These parts in turn divide the uvea into anterior (iris), intermediate (ciliary leather), and posterior (choroid).
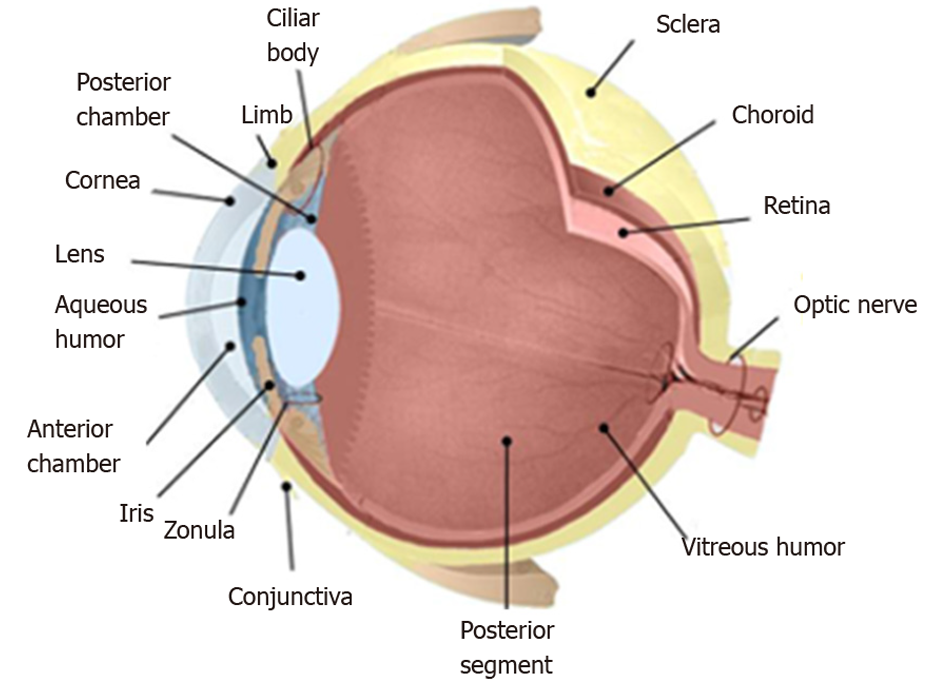
The anatomy of the eye is schematized in Figure 2, where the inside of the eye is represented. The drawing exhibits the interior of the eye including the lens, cornea, ciliary body, retina, choroid, vitreous humor, and optic nerve.
An ocular melanoma is a melanoma located in the eye or near the eye. This type of cancer develops in the cells that produce the pigment. The pigment is the substance that gives color to the eyes, skin, and hair. The melanoma develops in the skin, but it can also develop in the conjunctiva or in the eye.
The number of cancer deaths worldwide recorded by the World Health Organization is approximately 9.6 million in 2018. Therefore, cancer is the second leading cause of death globally. This data means that about one-sixth of all deaths are a result of cancer. Lung, prostate, colorectal, stomach and liver cancer are the most common cancers in the case of men. However, the most frequent types for women are breast, colorectal, lung, cervical and thyroid cancer.
Melanoma represents a small percentage among skin cancers, but it is responsible for the vast number of skin cancer deaths. Approximately half of patients with uveal melanoma develop metastases and die from the disease[1-4]. The most common site for metastatic spread is the liver. In most cases, liver metastases are due to a poor prognosis; and life expectancy is 4 to 15 mo if no treatment is performed[5,6]. The incidence of melanoma is increasing worldwide. It is estimated that in 2030 there will be a total of 23.6 million new cases[7].
Melanoma of the uvea is a rare kind of cancer, accounting for 5% of all melanomas[3,5,8,9]. Uveal melanoma represents the most widespread primary eye malignancy in adults, being exceptional in children, with an incidence of about 7 annual cases per million in Europe[10] and 5.3 to 10.9 annual cases per million in the United States[11]. In the case of Europe, incidence rates increase from South to North, being less than two cases per million in Southern Italy and Spain and more than 8 cases per million in Denmark and Norway[10]. In the United States, the difference in incidence among different ethnic groups is large, with the annual age-adjusted incidence being 0.38 for Asians, 0.31 for African Americans, 1.67 for Hispanics, and 6.02 for non-Hispanic whites[11]. However, the prognosis does not differ between ethnic groups[12].
The incidence increases with age, the maximum peak is reached between 60 and 70 years. It is slightly more frequent in men than in women and in people with iris and light skin. Normally, the affectation is unilateral and rarely bilateral. Solar exposure has been considered a possible contributing factor to the development of this tumor. It usually appears sporadically, although it is described as an increased factor in certain diseases: Uveal nevus, xeroderma pigmentosum, oculodermal melanocytosis (nevus of Ota), dysplastic nevus syndrome, and neurofibromatosis type I[13].
The incidence is variable depending on your location. Melanomas are more frequent in the choroid and less in the ciliary body and in the iris (90%, 7%, and 2% of uveal melanomas, respectively)[14]. The clinical and histopathological characteristics of conjunctival and uveal melanomas are distinct: The conjunctival is similar to cutaneous melanoma, and the uveal presents molecular similarities with melanocytic tumors of the central nervous system[15,16].
Mortality rates in five years are variable, between 6% and 53%, regardless of the first line treatment used for local control of the disease. It is known that approximately 50% of patients will develop distant metastasis, mainly in the liver, lung, bone, and skin[17,18]. The average survival period after diagnosis is about seven months.
At the present, there is no treatment for metastatic uveal melanoma. The survival rates have remained stable since the 1970s despite advances in treatment and knowledge of tumor biology. In this sense, it is essential to deepen the knowledge of the molecular actors involved in the initiation and progression of the tumor.
Despite the complexity of the mechanisms of cell biology, we can analyze them in depth by means of their structure, dynamics and control procedures. Predictive models can provide a great benefit for the knowledge of signaling pathways processes in humans. Basically, these molecular pathways carry out the detection of cells, transformation/modification of their components, and transmission of information from their environment to intracellular targets[19,20].
There are numerous perspectives for computational analysis of cellular signaling networks, such as statecharts[21], ordinary differential equations[22], Petri nets[23,24], live sequence charts[25], and ambient/membrane calculi[26], and rule-based[27,28]. Quantitative analyses require the use of a large number of molecules per species. However, in the case of huge numbers, the complexity increases enormously. Qualitative modeling supplies alternative approaches when quantitative methods do not give efficient solutions.
Computational analysis with qualitative approaches have provided a breakthrough in research cell biology and medical biology[29-32]. Symbolic models allow us to model, compute, analyze, and reason about networks of molecular interactions at multiple levels of detail[33,34]. Such models can suggest new knowledge and understanding of complex biological processes. This formalism provides a language for representing system states (different elements, with their locations that are present in the cell at a given time) and mechanisms of change (such as reactions), as well as analysis tools based on logical inference. In this way, behavior of a system can be simulated by symbolic models. The goal is to achieve formal models that are closer to the mindset of biologists[35].
Rule-based models allow managing biological interactions in a natural manner[36-38]. Highly complex cellular processes are successfully and efficiently handled due to the competence of rule-based systems which deal with complex systems[28,39-43].
The rest of the paper reviews the main features of uveal melanoma in Section 2. A description of signaling pathways involved in uveal melanoma is presented in Section 3. The application of artificial intelligence (AI) in modeling and analysis of signaling pathways involved in uveal melanoma with rule-based symbolic systems is presented in Section 4. Finally, we draw our discussion in Section 5.
The etiology of uveal melanoma is not yet clear[44]. Ultraviolet radiation (UV radiation) is established as the main risk factor for cutaneous melanoma, although the role of UV radiation in the development of uveal melanoma remains controversial[45,46]. On the other hand, we also comment on the possible influence of genetic factors and somatic mutations.
Ultraviolet radiation: Population pigmentation and geographical parameters, such as latitude and altitude, influence the incidence of melanoma. This indicates that UV radiation has a causal role in the development of melanoma[47,48]. The solar radiation that reaches the earth’s surface is a range of electromagnetic radiation that is composed of two ranges of ultraviolet wave bands: 95% ultraviolet A (with range between 320 and 400 nm) and 5% ultraviolet B (with range between 280 and 230 nm). The role of these two types of waves is different in the ability to initiate DNA damage, cell signaling pathways and immune alterations[7,49].
Ultraviolet B is considered the main carcinogen of melanoma[50]. The predominant photo-lesions induced by ultraviolet B are: the DNA cyclobutane pyrimidine dimers, pyrimidine-6, 4-pyrimidone photoproducts, and Dewar photoproducts. These DNA lesions are only repaired by a nucleotide excision repair system. If unrepaired, these mutations at dipyrimidine sites induce the characteristic ultraviolet-signature mutation[7]. On the other hand, ultraviolet A wavelengths interact with cellular photosensitizers to produce reactive oxygen species and oxidative damage to DNA. Although rare, ultraviolet radiation also has the capacity to induce other types of DNA alterations, such as protein-DNA crosslinks, single-strand breaks, oxidative base damage, epigenetic changes, and chromosomal aberrations[7].
Exposure to ultraviolet radiation produces numerous cellular reactions, such as epidermal hyperplasia, cutaneous inflammation, and migration of melanocyte stem cells to the interfollicular epidermis. It has been observed that melanoma occurs commonly after intermittent sun exposure and in people with frequent sunburns, especially during childhood[51]. Risk of melanoma has also been associated with high-dose use of indoor artificial tanning devices[52]. However, chronic or low-grade exposures to ultraviolet radiation induce DNA protection due to increased skin thickness and melanin production resulting from chronic ultraviolet exposure[51,53].
Genetic factors: A common characteristic of melanoma patients is a pale-skinned complexion, red or blond hair, blue eyes and a high number of large and irregular nevi. The presence of nevi has a high correlation with exposure to ultraviolet radiation. Familial melanomas constitute 8%–12% of all melanoma cases and allow identification of the melanoma susceptibility genes involved in the familial disease, even in sporadic cases[7,54].
Somatic mutations: The interruption of the precise control of the transduction of cell signaling pathways is linked to many oncogenes and tumor suppressors. Signal transduction is the complex communication system that coordinates the actions of cells and governs cellular activities[55]. Poor regulation of this network can induce the acquisition of cancer phenotypes. In this way, cell signaling pathways allow us to understand the processes that are also closely involved in cancer: Cell growth and death, migration, metabolism, angiogenesis, and so on.
Uveal melanoma can develop without any symptoms and is diagnosed by a routine eye examination. It often causes painless distortion of vision and other nonspecific visual symptoms[56].
The diagnosis of uveal melanoma consists of clinical examination and ocular ultrasonography. The high levels of accuracy and detection rates at the first visit of an experienced eye oncologist make it possible to avoid an invasive diagnostic biopsy[57,58]. Delayed operating time may affect prognosis, especially in older patients with smaller tumors.
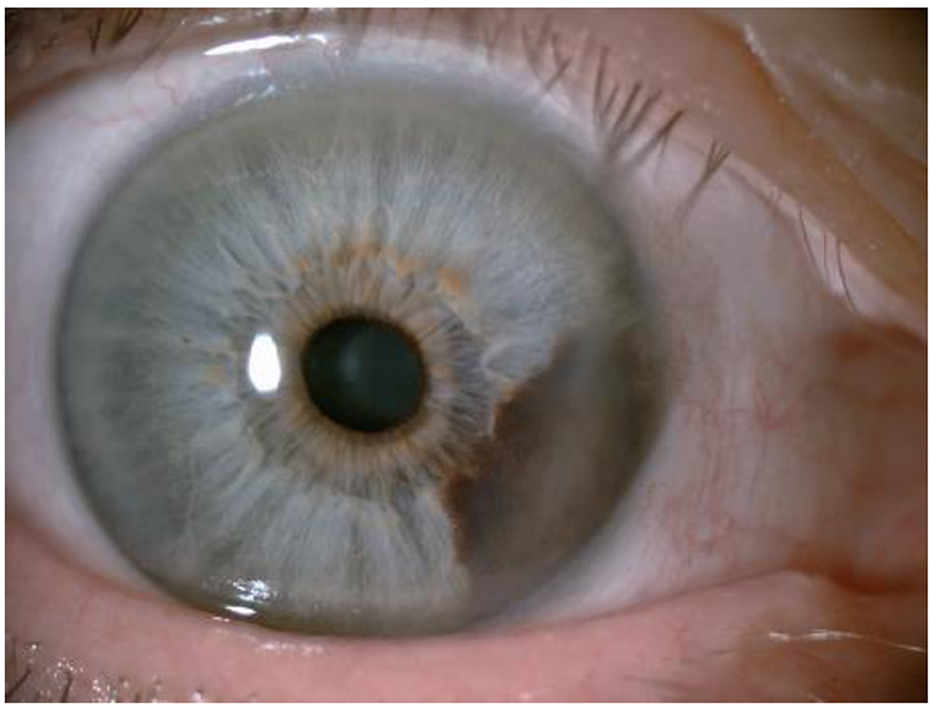
For melanoma of the anterior uvea (iris), the best diagnostic criteria are evidence that the lesion is growing. Figure 3 shows an iris melanoma that extends to the ciliary body.
For the diagnosis of posterior uvea melanoma (ciliary body and choroids), transillumination and fundoscopic examination through pupil dilation (indirect ophthalmoscopy) remain the first steps in the diagnostic process[59]. A choroidal nevus in a patient with choroidal melanoma of the contralateral eye is shown in Figure 4.
A shortcoming is the diagnosis of small tumors since it is not possible to distinguish whether they are melanomas or nevi. For this purpose, specialized ocular imaging techniques are useful in order to detect clinical signs that can help in the differential diagnosis, such as: thickness > 2 mm, subretinal fluid, symptoms related to alteration of vision, orange pigmentation, tumor very close to the optic nerve, absence of dorsum, absence of acoustic halo in the US, absence of pattern with pigmented halo, which favor the diagnosis of malignancy[60,61].
For the extension study, in order to assess metastatic lesions, the techniques used are: Computerized tomography of the chest and abdomen, positron emission tomography PET-CT, ultrasonography, and abdominal magnetic resonance imaging (liver)[61]. The usefulness of detecting circulating tumor cells in the bloodstream in order to discover patients at risk of metastasis is questioned[62]. In oncology, the treatment of malignant tumors usually requires histological confirmation of the clinical diagnosis. For intraocular melanoma, therapeutic decisions are mostly based on clinical diagnosis[63].
There are different ways of treatment: (1) Surgery (local resection, endoresection, or enucleation); and (2) Local radiotherapy (106-ruthenium or 125-iodine brachytherapy, proton beam therapy, or stereotactic radiosurgery)[58]. To minimize the side effects of brachytherapy, the neoadjuvant phototherapy is proposed[64].
Currently, the management of posterior uveal melanoma depends on several factors such as: size of the tumor, extension, age of the patient, general health status, condition of the opposite eye, patient’s desire and psychological status.
After proton beam therapy, local control of the disease is achieved in 96.4%[65], however local recurrence can take place up to almost ten years after primary therapy and poses a higher risk of metastasis[66].
The malignant potential of tumors has been a great concern for years. Therefore, it is important to understand why some patients evolve more torpidly and quickly than others despite having the same type of neoplasm. For this reason, we need to be able to recognize factors intrinsic to the tumor or to the patient himself, that allow classifying and/or predicting the evolution of the course of the disease (prognostic factors) in order to be able to offer effective and/or preventive treatments. In uveal melanoma, several clinical, histopathological, cytogenetic and molecular factors have been described which allow the identification of those patients who present a higher risk of developing distant metastasis and who could probably benefit from an adequate prophylactic and/or adjuvant treatment[59,67].
Some features increase the likelihood of developing uveal melanoma. Age, sex, and ethnicity are related to different incidence rates of the disease. The risk of uveal melanoma increases with age (the peak value is reached at age 70), men develop it more often than women and Caucasians are more likely to develop it compared to populations with a darker skin type. In the development of uveal melanoma, there are some other risk factors such as fair skin, inability to tan, light eye color, and blond hair. Other clinical features associated with an increased risk of uveal melanoma are oculodermal melanocytosis and cutaneous, iris and choroidal nevi[5,68].
According to the literature[59], the prognostic factors that have been described in uveal melanoma are: (1) Clinical: Age and sex, location and configuration of the tumor, tumor size and staging [American Joint Committee on Cancer develops a classification system for describing the extent of disease progression in cancer patients (https://cancerstaging.org/)], association with ocular or oculodermal melanocytosis; (2) Histological: Cellular type and nuclear size, mytotic activity, vascular pattern and density, inflammatory infiltrate, necrosis and pigmentation; and (3) Cytogenetic and molecular: cytogenetic alterations and molecular alterations.
The interruption of the precise control of the transduction of the cell signaling pathways is related to several oncogenes and tumor suppressors. Management of cellular activities and coordination of the actions of the cells constitute a complex system of communication known as signal transduction. A poor regulation of this network can lead to the acquisition of cancer phenotypes. Cellular signaling pathways are also fundamental to understanding processes that are also closely related to cancer: cell growth and death, migration, metabolism, and angiogenesis.
From the genetic point of view, melanoma is a complex disease[69]. Its genetic alterations affect genes in key signaling pathways that govern: (1) Proliferation (NRAS, BRAF, and NF1); (2) Growth and metabolism (STK11, PTEN, and KIT); (3) Replicative response (TERT); (4) Cell cycle control (CDKN2A); and (5) Resistance to apoptosis (TP53).
Knowledge of the molecular system of uveal melanoma has improved in recent decades and is constantly being updated. Unlike cutaneous melanoma, in uveal melanoma, NRAS, BRAF, NF1, and c-KIT mutations are rarely produced. However, some other mutations have been detected: BAP1, EIF1AX, CYSLTR2, GNA11, GNAQ, PLCβ4, and SF3B1. These genetic changes allow us to better categorize patients according to the individual risk of distant metastasis[70]. The most common mutations identified in primary uveal melanoma are GNAQ/11 mutations. GNAQ mutations are present in up to half of the cases[71,72].
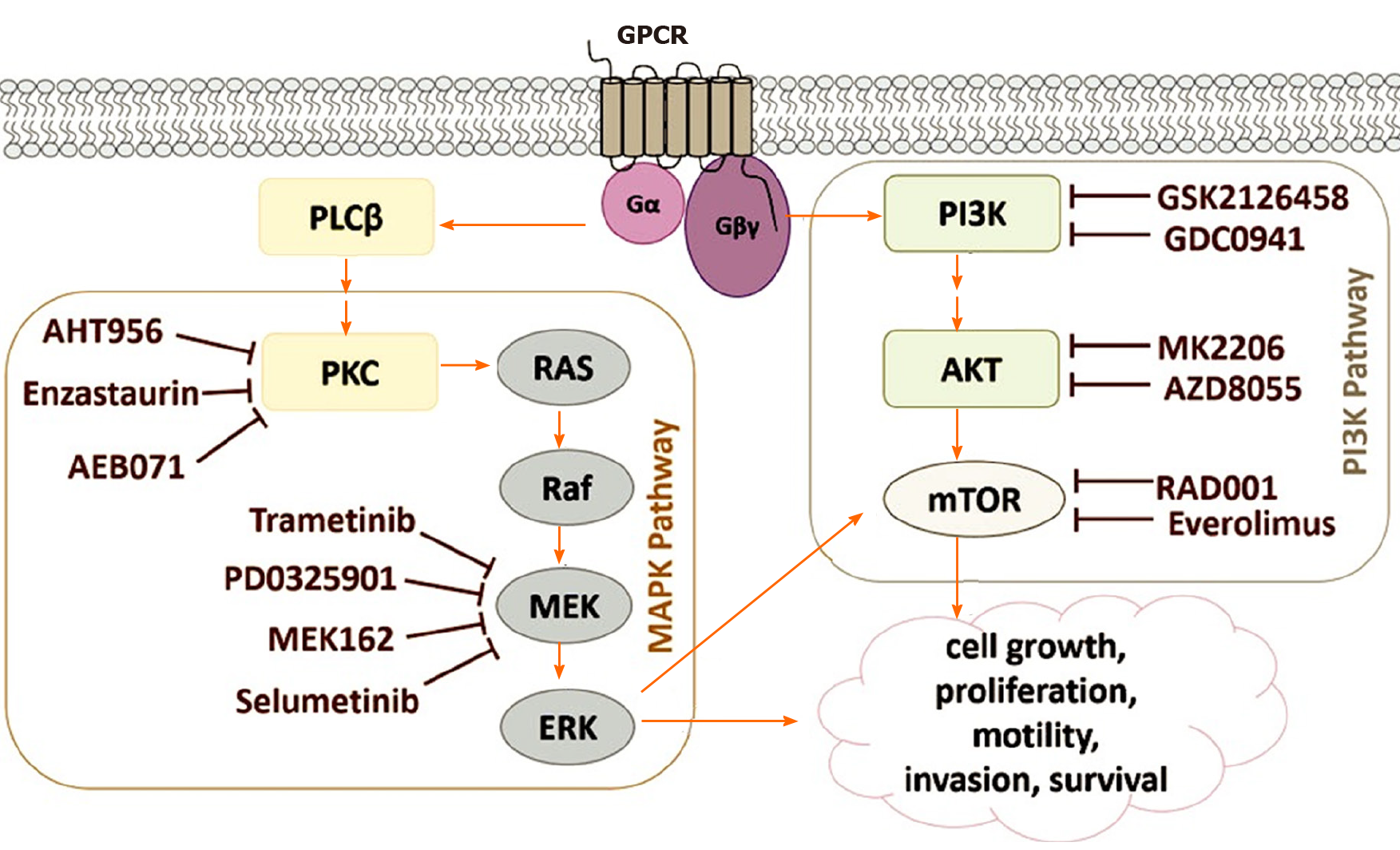
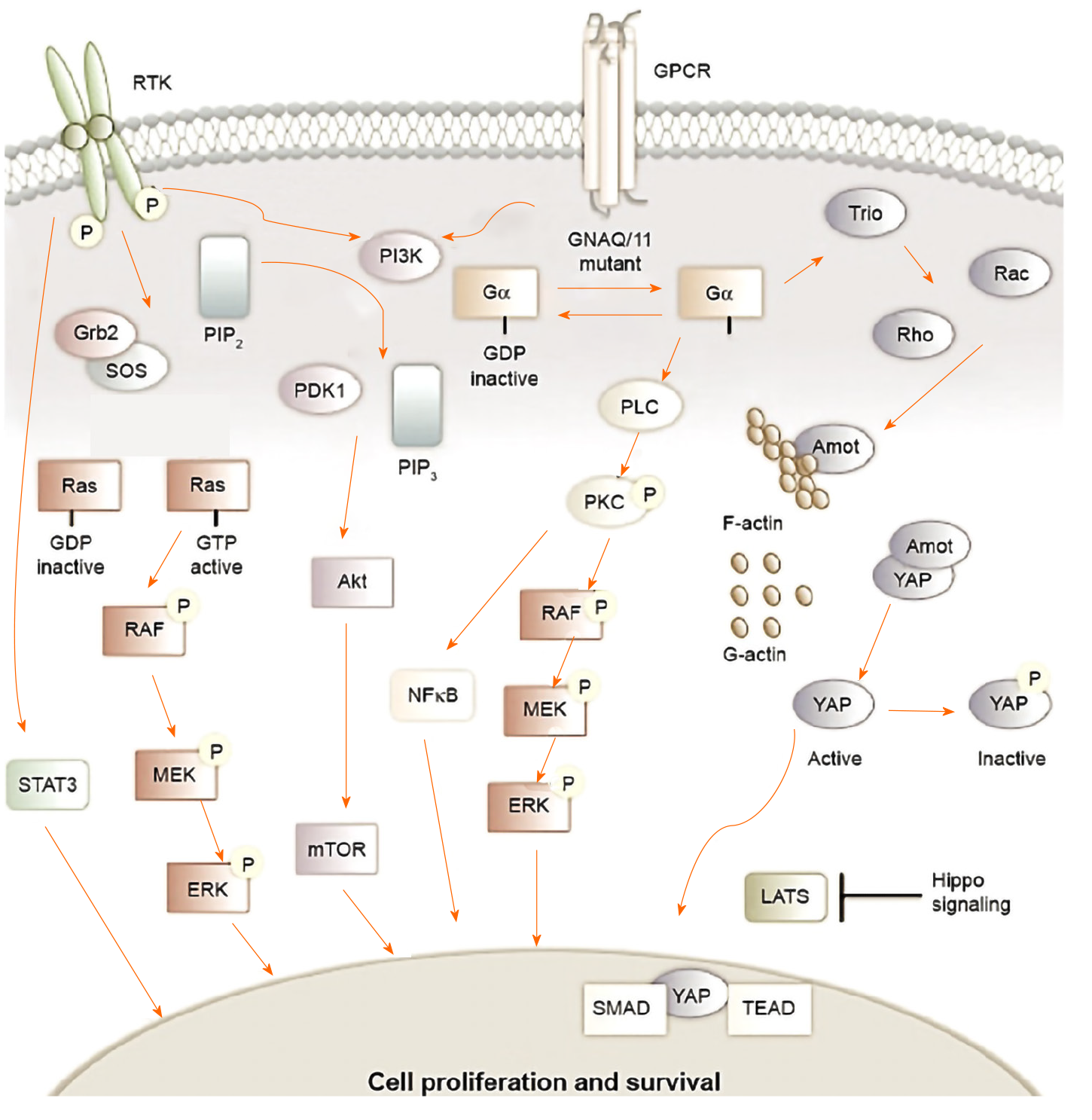
GNAQ/11 mutations that occur in uveal melanomas cause PI3K/Akt/mTOR signaling to be regulated upwards. The activation of the PI3K pathway triggers antiapoptotic signals that complement the proliferative effects of the overactive MAPK signaling that contributes to uveal melanomagenesis[5,73]. Figure 5 shows an outline of the signaling pathways involved in the development of uveal melanoma, as well as the specific inhibitors used in preclinical studies (based on Álvarez-Rodríguez et al[5]).
The activation of GPCR, a type of receptor on the cell surface, can be caused by various stimuli. This type of activation triggers the exchange of GDP to GTP in the Gα subunit of the heterotrimeric G-protein leading to the dissociation of the Gα subunit from the other two Gβγ subunits. Some mutations can produce activation of G-proteins, such as those that occur in GNAQ or GNA11 in 75% of uveal melanomas. After activation, the subunit Gα causes the cleavage of PIP2 into IP3, and DAG by PLCβ. The activation of the MAPK pathway is followed by the activation of PKC (protein kinase C) by DAG. In addition, Gβγ activates the PI3K route. These two pathways are involved in cell survival, mobility, growth, invasion, and proliferation[5].
Complex biological mechanisms can be thoroughly revealed by their structure, dynamics, and control methods. AI helps the research of human signaling processes[74,75]. Molecular pathways detect cells, transform components, and internally transmit information from their environment to intracellular targets, such as the genome[20,76].
Symbolic models can be used to model, compute with, analyze, and reason on nets of molecular interactions at different levels of detail, depending on the information available and the aspects to be investigated. These models facilitate a deep knowledge of biological processes and new relationships between their elements. This formalism includes a language to model the states of the system and the dynamics of change (such as reactions), as well as computational inference or logic tools that allow us to analyze the processes[75,77]. In this manner, system behavior can be mimicked with symbolic models. The objective is to establish formal models close to the mentality of biologists[35]. Rule-based models can handle molecular interactions in a natural way. The skills of rule-based systems allow to deal with schemes of a large underlying complexity. In this way, complex cellular processes are managed adequately and efficiently[28].
We define a rule-based system which allows us to carry out an analysis of the change of different initial states, and of the study of the states that can be reached at the initial states in the signaling pathways involved in uveal melanoma. This task is carried out thanks to rewriting logic and Pathway Logic, which we briefly describe below.
Rewriting logic and Pathway Logic: Rewriting logic constitutes a logic of change or becoming[78]. It allows you to easily set the specification of the dynamic features of systems and naturally deals with highly nondeterministic concurrent computations. Rewriting logic provides a flexible and general semantic framework to confer semantics to a wide range of languages and concurrency models[79]. Rewriting logic is efficiently implemented in the Maude language[80].
On the one hand, the rewriting logic consists of an equational theory that define sorts, constructors, function symbols, and equality between terms. On the other hand, the rewriting logic extends the equational theory with rewrite rules that allows expressing the dynamics between the states of the system. Rewrite rules lay down local and parallel changes in a dynamic concurrent system. In this manner, these deduction rules establish a sound reasoning. From a purely logical point of view, we will say that each rewrite rule is a logical entailment in a formal model.
Based on rewriting logic, Pathway Logic[27] is a platform for modeling and analyzing molecular and cellular processes. The resulting formal models can be executed and analyzed using the Maude system[81]. Many models have been developed with Pathway Logic because of the naturalness of rewriting logic to model and experiment with mathematical and biological problems[82,83]. Pathway Logic is presently being used to curate several models of signal transduction and metabolic networks[31].
A rule knowledge base in Pathway Logic consists of rewrite rules and supporting data type specifications[27]. The model of melanoma signaling system consists of: (1) A specification of the starting cell components with their locations, the so-called initial state; and (2) A collection of rewrite rules derived from the global knowledge base by a symbolic reasoning process that recruits all rewrite rules that are potentially executable from the initial state. These executable models collect the possible paths in which a system can progress. Logical inference of Pathway Logic can: (1) Simulate possible ways in which a system could evolve; (2) Build pathways in response to queries; and (3) Think logically about dynamic assembly of complexes and cascade transmission of signals[19,84].
Modeling of signaling pathways in uveal melanoma: Through the language Maude and pathway logic, the various elements found in a cell (proteins, genes, chemicals, etc.) are defined as a Soup (i.e. a set or an associative and commutative list with a neutral element). Such elements constitute a location and are identified by a location name (LocName): op {_|_}: LocName Soup -> Location [ctor].
Some of the various parts or locations of the cell can be: In the nucleus (NUc), in the cytoplasm (CLc), in/across the cell membrane (CLm), outside the cell (XOut), or attached to the inside of the cell membrane (CLi).
In the following code fragment, the nucleus location (NUc) is defined with some elements, such as genes and proteins (e.g., Maz, Myc, and Rb1), some of which are modified (e.g., a high mRNA expression level of Tp53 gene is presented: [Tp53-gene - on]): {NUc | Maz Myc Rb1 NProteasome Chek2 Chek1 Tp53 [Tp53-gene - on]}.
In turn, we can have a set or Soup of the different locations of the cell with their corresponding contents. At last, all location sets (Soups) are collected in wrappers called dishes, through the PD operator.
For the purpose of better understanding the modeling of a state, a small dummy cell is represented in Maude with the following dish:
op DummyDish: -> Dish.
eq DummyDish = PD({CLm | Erbb2 Igf1R [Cbl - Yphos]} {XOut | Igf1 } {CLi | [Gnai1 - act] [Hras - GDP]} {NUc | Elk1 Msk1 Maz} {CLc | Mek1 Akts [Csnk1a1 - act] [Gsk3s - act]}).
Several proteins are included in the dish, such as receptor tyrosine-protein kinase erbB-2 (written as Erbb2 according to Pathway Logic notation), insulin-like growth factor I (Igf1), and Myc-associated zinc finger protein (Maz). Some of these components have modifications, such as phosphorylation on tyrosine (Yphos), binding to GDP, or activation (act). On the other hand, the ligand/receiver bond between cell components can be defined in Maude with the operator (_:_). As an example, a bond between Egf and EgfR is written in Maude as (Egf : EgfR). A pictorial and informal representation of this dummy cell is shown in Figure 6. In this figure, unmodified proteins are shown in green. Proteins with modifications are exhibited with different colors: red for activated proteins, blue for phosphorylated proteins, and yellow for those bound to GDP.
Below, we model a complete dish MELANOMADish for a melanoma case study. This cellular dish is composed of several locations, such as CLc or NUc. The contents of each cell location, such as CLc or NUc, are defined as a soup of elements. Each of the elements or components, such as Akts, may include some modifications (e.g., [Rheb - GTP], [Gsk3s - act], etc.). Here is a rough version of Maude’s module containing this dish:
mod MELANOMA is inc ALLOPS.
op MELANOMADish : -> Dish.
eq MELANOMADish = PD(
{CLc | [Csnk1a1 - act] [Gsk3s - act] [Ilk - act] Akts Igf1R Axin1 BrafV600E Btrc Rnf6 Trim28 Bim Ctnnb1 Cul7 Eif4ebp1 Pkca Erks Cdc42 Fbxw8 Irs1 Cdkn2a Mek1 Mlst8 Mtor Pdpk1 Erbb2 Ep300 Proteasome Rac1 Rad54b Rbbp6 Raptor Rbx1 Rps6 Rictor Mdm4 Ube2d2 Ube2d1 Rsk1 S6k1 Sin1 Skp1 Ybx1 Ywhas Akt1 Ang Dzip3 Baiap2 L1 C10orf90 Cdk5rap3 Cbp Cul5 G3bp1 Rchy1 Rnf31 Syvn1 Magea3 Erk5 Mdm2 Mkrn1 NgfR Nr0b2 Nus1 Pax3 Pcmt1 Pdlim7 Pkch Ppm1d Psme3 Ube4b Rnf43 Stub1 Tax1bp3 Huwe1 Gli1 Tpt1 Trim24 Ube2d3 Ubc13 Ube3a}
{NUc | [Tp53-gene - on] Maz Myc Rb1 Chek1 NProteasome Chek2 Tp53}
{CVc | [Rheb - GTP] (Tsc1 : Tsc2)} {Sig | empty})
{CLm | PIP2} {XOut | empty} {CLi | Pld1 Pi3k Parva} {CLo | empty}.
endm
Rule-based dynamics in uveal melanoma: According to the literature, the PI3K, MAPK, IGF-1R, and mTOR pathways are actively involved in uveal melanoma[85-87]. Based on Krantz et al[85], Figure 7 illustrates the main signaling pathways that influence uveal melanoma.
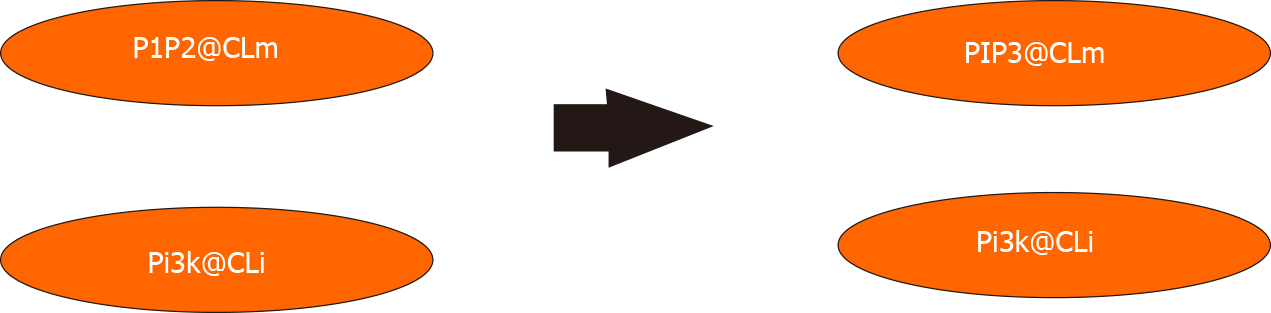
Maude’s rewriting rules establish the dynamics of our biological system. Concurrent cellular reactions can be defined with these rules. To illustrate this, the 3820c rule states that Pi3k the inside of the cell membrane mediates phosphorylation of PIP2 into PIP3 in the cell membrane [The variables clm and cli indicate that they can be replaced by any soup of elements in/across the cell membrane and attached to the inside of the cell membrane, respectively (Figure 8)]: rl[3820c.PIP3.from.PIP2]: {CLi | cli Pi3k} {CLm | clm PIP2} => {CLi | cli Pi3k} {CLm | clm PIP3}.
Each of these rules is extracted from scientific knowledge. In the case of rule 3820c, the evidence was obtained from KEGG and HumanCyc curated databases containing metabolic reactions [HumanCyc reaction 2.7.1.153 (http://humancyc.org/HUMAN/ NEW-IMAGE?type=NIL&object=2.7.1.153-RXN), and KEGG reaction R04545 (http://www.kegg.jp/dbget-bin/www_bget?rn:R04545)].
Once the biological system has been modeled with the elements involved (proteins, genes, etc.) and with the rewriting rules that define the dynamics of the model, we can now express the potential of rewriting logic and the environment of Pathway Logic to analyze our biological system from different points of view and draw inferences.
For example, the rewrite command allows to apply rewrite rules and obtain a reachable dish from our initial dish. That is, starting from the cell that we have defined with MELANOMADish, we obtain the final state of the cell only after five of the possible reactions have taken place. The result of applying five rewrite steps to our initial dish is shown in the following example:
Maude > rewrite [5] MELANOMADish.
result Dish: PD({CLm | PIP3} {CLo | empty} {CLi | Pld1 Pi3k Parva}
{NUc | NProteasome Rb1 Tp53 Maz Chek1 Myc Chek2 [Tp53-gene - on]}
{CVc | [Rheb - GTP] (Tsc1 : Tsc2)} {XOut | empty} {Sig | empty}
{CLc | Ube2d3 Erks Fbxw8 Ybx1 G3bp1 Ang Ywhas Ctnnb1 Cul7 Cul5 Axin1 Bim Btrc Mdm4 BrafV600E NgfR Mtor Cdkn2a C10orf90 Pdlim7 Pkch Eif4ebp1 S6k1 Sin1 Syvn1 Tax1bp3 Pkca Proteasome Ppm1d Dzip3 Rictor Rnf6 Rnf31 Rnf43 Erbb2 Ep300 Erk5 Gli1 Trim28 Baiap2 L1 Ubc13 Huwe1 Ube2d2 Mdm2 Magea3 Mek1 Stub1 Mlst8 Pax3 Pcmt1 Psme3 Rac1 Igf1R Irs1 Rbbp6 Rad54b Raptor Ube3a Cbp Cdc42 Mkrn1 Ube4b Cdk5rap3 Rbx1 Nus1 Nr0b2 Rchy1 Rps6 Rsk1 Skp1 Tpt1 Trim24 Akts Akt1 [Gsk3s - act] [Ilk - act] [Csnk1a1 - act] [Pdpk1 - act] Ube2d1 })
In this first possible solution, some activated proteins are observed in the cytoplasm, such as Gsk3s, Csnk1a1, and Ilk. However, the possible results of rewriting a term may be different, depending on the rules and the order in which they are applied.
Moreover, we can also carry out a breadth-first search with a given pattern using the search instruction. In the following example, we are looking for two states of a cell that satisfy the following conditions: (1) An Erks protein is activated in the nucleus or cytoplasm; (2) A Pi3k protein is attached to the inside of the plasma membrane; and (3) Each of the cell states is reached in a maximum of five steps.
search [2,5] MELANOMADish =>* PD(S:Soup {NUc | nuc:Things}
{CLi | cli:Things Pi3k} {CLm | clm:Things PIP3}
{loc:LocName | things:Things [Erks - erksmodset:ModSet act]})
such that (loc:LocName == NUc) or (loc:LocName == CLc).
In this example, we use the search option =>*, which means that the search must be performed in zero or more steps. Moreover, the variable S:Soup in the search pattern indicates the rest of elements. Maude achieves two possible solutions which fulfill these conditions and displays the terms that show matching/adjustment in the solutions.
Solution 1 (state 186)
S:Soup --> {CLo | empty} {CVc | [Rheb - GTP] (Tsc1 : Tsc2)} {Sig | empty}
{XOut | empty}
nuc:Things --> Maz Myc Chek1 NProteasome Rb1 [Tp53-gene - on] Chek2 Tp53
clm:Things --> empty cli:Things --> Parva Pld1
loc:LocName --> CLc erksmodset:ModSet --> phos(TEY)
things : Things --> C10orf90 Cdk5rap3 Rnf43 Rsk1 Cdkn2a Cul7 Dzip3 Erbb2 Magea3 Huwe1 Stub1 Syvn1 Mkrn1 Mtor Cdc42 NgfR Nr0b2 Pcmt1 Ube2d1 Ube2d2 Pdpk1 Pkca Eif4ebp1 Rps6 Ep300 Pkch Ppm1d G3bp1 Gli1 Psme3 Fbxw8 Proteasome Rac1 Mdm2 Mlst8 Rad54b Cbp Raptor Rbbp6 Nus1 Pax3 Rbx1 Rchy1 Akts Ube2d3 Ube3a Pdlim7 Akt1 Ang Axin1 Baiap2 L1 Bim [ Csnk1a1 - act] [Gsk3s - act] Btrc Rictor [Mek1 - act phos(SMANS)] Rnf6 Rnf31S6k1 Igf1R Sin1 Mdm4 Skp1 Tax1bp3 Tpt1 Ybx1 Trim24 Erk5 Irs1 Trim28 Ubc13 Ube4b Ctnnb1 Cul5 Ywhas [Braf - act] [Ilk - act]
The set of modifications erksmodset:ModSet of this solution contains the protein Erks which is activated and phosphorylated in TEY domain.
In addition, the rules that have been applied to reach state 186 can be obtained by using the instruction show path labels:
Maude > show path labels 186.
3820c.PIP3.from.PIP2
3808c.BrafV600E.act
431c.Mek1.by.Braf
014c.ErkS.by.Mek1
The output of the previous command indicates that 3820c, 3808c, 431c, and 014c rules have been applied to the initial state (MELANOMADish).
In 2018 the World Health Organization recorded approximately 9.6 million cancer deaths worldwide. Therefore, cancer is the second leading cause of death. This data means that about one-sixth of all deaths are the result of cancer. Melanoma represents only about 1% of skin cancer, but it is responsible for the vast majority of skin cancer deaths. Uveal melanoma is a rare type of cancer and represents up to 5% of all melanomas[3,5,8,9]. Approximately half of patients with uveal melanoma develop metastases and die from the disease[1-3].
Symbolic systems biology can explore and analyze biochemical reactions that occur concurrently in a cell. The use of rewriting rules of AI models allows the modeling of biological processes in the cell[74,77]. Our final goal is to provide models that encloses the reasoning and intuitions of biologists.
The computational analyses with qualitative approaches have brought about a breakthrough in research in medical biology and cell biology[19,20]. Symbolic models allow us to model, compute, analyze and reason on networks of molecular interactions at multiple levels of detail[33,34]. Such models can suggest new knowledge and understanding of challenging cellular processes. This formalism provides us with a language that can to be able to represent system states and mechanisms of change and with tools to perform logical inferences and other meta-analyses[32,81].
This paper gives an overview of the computational analysis of signaling pathways based on rewriting logic paradigm. Pathway Logic's SKMELL33 model provides a specific symbolic logic system that browses the complex and dynamic cellular signaling processes that lead to cell survival and proliferation in uveal melanoma[27,84]. The understanding of the signaling pathways involved in melanoma will offer new strategies for effective treatments.
The authors are highly grateful to anonymous reviewers for their valuable comments and helpful suggestions.
Manuscript source: Invited manuscript
Specialty type: Cell Biology
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Youness RA S-Editor: Wang JL L-Editor: A P-Editor: Ma YJ
| 1. | Carvajal RD, Schwartz GK, Tezel T, Marr B, Francis JH, Nathan PD. Metastatic disease from uveal melanoma: treatment options and future prospects. Br J Ophthalmol. 2017;101:38-44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 251] [Cited by in RCA: 288] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 2. | Char DH. Metastatic choroidal melanoma. Am J Ophthalmol. 1978;86:76-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 87] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 3. | Chattopadhyay C, Kim DW, Gombos DS, Oba J, Qin Y, Williams MD, Esmaeli B, Grimm EA, Wargo JA, Woodman SE, Patel SP. Uveal melanoma: From diagnosis to treatment and the science in between. Cancer. 2016;122:2299-2312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 286] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 4. | Karlsson J, Nilsson LM, Mitra S, Alsén S, Shelke GV, Sah VR, Forsberg EMV, Stierner U, All-Eriksson C, Einarsdottir B, Jespersen H, Ny L, Lindnér P, Larsson E, Olofsson Bagge R, Nilsson JA. Molecular profiling of driver events in metastatic uveal melanoma. Nat Commun. 2020;11:1894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 5. | Álvarez-Rodríguez B, Latorre A, Posch C, Somoza Á. Recent advances in uveal melanoma treatment. Med Res Rev. 2017;37:1350-1372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 6. | Dogrusöz M, Jager MJ, Damato B. Uveal Melanoma Treatment and Prognostication. Asia Pac J Ophthalmol (Phila). 2017;6:186-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 7. | González Sánchez E. Study of the cooperative effect of UVR, BRAF and LKB1 in melanoma. PhD thesis, Autonomous University of Barcelona. 2019. Available from: https://hdl.handle.net/10803/667375. |
| 8. | Egan KM, Seddon JM, Glynn RJ, Gragoudas ES, Albert DM. Epidemiologic aspects of uveal melanoma. Surv Ophthalmol. 1988;32:239-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 339] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 9. | Kaliki S, Shields CL. Uveal melanoma: relatively rare but deadly cancer. Eye (Lond). 2017;31:241-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 425] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 10. | Virgili G, Gatta G, Ciccolallo L, Capocaccia R, Biggeri A, Crocetti E, Lutz JM, Paci E; EUROCARE Working Group. Incidence of uveal melanoma in Europe. Ophthalmology. 2007;114:2309-2315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 305] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 11. | Singh AD, Topham A. Incidence of uveal melanoma in the United States: 1973-1997. Ophthalmology. 2003;110:956-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 333] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 12. | Shields CL, Kaliki S, Cohen MN, Shields PW, Furuta M, Shields JA. Prognosis of uveal melanoma based on race in 8100 patients: The 2015 Doyne Lecture. Eye (Lond). 2015;29:1027-1035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 96] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 13. | Laver NV, McLaughlin ME, Duker JS. Ocular melanoma. Arch Pathol Lab Med. 2010;134:1778-1784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 14. | Shields CL, Manalac J, Das C, Ferguson K, Shields JA. Choroidal melanoma: clinical features, classification, and top 10 pseudomelanomas. Curr Opin Ophthalmol. 2014;25:177-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 15. | Amaro A, Gangemi R, Piaggio F, Angelini G, Barisione G, Ferrini S, Pfeffer U. The biology of uveal melanoma. Cancer Metastasis Rev. 2017;36:109-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 149] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 16. | Smit KN, Jager MJ, de Klein A, Kiliҫ E. Uveal melanoma: Towards a molecular understanding. Prog Retin Eye Res. 2020;75:100800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 181] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 17. | Gragoudas ES, Egan KM, Seddon JM, Glynn RJ, Walsh SM, Finn SM, Munzenrider JE, Spar MD. Survival of patients with metastases from uveal melanoma. Ophthalmology. 1991;98:383-9; discussion 390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 240] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 18. | Woodman SE. Metastatic uveal melanoma: biology and emerging treatments. Cancer J. 2012;18:148-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 19. | Santos-Buitrago B, Riesco A, Knapp M, Santos-García G, Talcott CL. Reverse inference in symbolic systems biology. In: Fdez-Riverola F, Mohamad MS, Rocha MP, De Paz JF, Pinto T, editors. Volume 616 of Advances in Intelligent Systems and Computing. 11th International Conference on Practical Applications of Computational Biology & Bioinformatics, PACBB 2017; 2017 June 21-23; Porto, Portugal. Springer, 2017: 101-109. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 20. | Weng G, Bhalla US, Iyengar R. Complexity in biological signaling systems. Science. 1999;284:92-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 460] [Cited by in RCA: 359] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 21. | Efroni S, Harel D, Cohen IR. Toward rigorous comprehension of biological complexity: modeling, execution, and visualization of thymic T-cell maturation. Genome Res. 2003;13:2485-2497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 69] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 22. | Stei MM, Loeffler KU, Holz FG, Herwig MC. Animal Models of Uveal Melanoma: Methods, Applicability, and Limitations. Biomed Res Int. 2016;2016:4521807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 23. | Hardy S, Robillard PN. Petri net-based method for the analysis of the dynamics of signal propagation in signaling pathways. Bioinformatics. 2008;24:209-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 24. | Li C, Ge QW, Nakata M, Matsuno H, Miyano S. Modelling and simulation of signal transductions in an apoptosis pathway by using timed Petri nets. J Biosci. 2007;32:113-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Sadot A, Fisher J, Barak D, Admanit Y, Stern MJ, Hubbard EJ, Harel D. Toward verified biological models. IEEE/ACM Trans Comput Biol Bioinform. 2008;5:223-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 17] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Regev A, Panina EM, Silverman W, Cardelli L, Shapiro E. BioAmbients: An abstraction for biological compartments. Theor Comput Sci. 2004;325:141–167. [DOI] [Full Text] |
| 27. | Talcott CL. Pathway Logic. In: Bernardo M, Degano P, Zavattaro G, editors. Advanced Lectures, volume 5016 of Lecture Notes in Computer Science. Formal Methods for Computational Systems Biology 8th International School on Formal Methods for the Design of Computer, Communication, and Software Systems, SFM 2008; 2008 Jun 2-7; Bertinoro, Italy. Springer, 2008: 21-53. [DOI] [Full Text] |
| 28. | Hwang W, Hwang Y, Lee S, Lee D. Rule-based multi-scale simulation for drug effect pathway analysis. BMC Med Inform Decis Mak. 2013;13 Suppl 1:S4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 29. | Burnier JV, Mastromonaco C, Lasiste JM, Burnier MN. Animal models in uveal melanoma. In: Uveal Tumors, Clinical Ophthalmic Oncology. Cham: Springer, 2019: 135-154. [DOI] [Full Text] |
| 30. | Metzcar J, Wang Y, Heiland R, Macklin P. A Review of Cell-Based Computational Modeling in Cancer Biology. JCO Clin Cancer Inform. 2019;3:1-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 237] [Cited by in RCA: 206] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 31. | Riesco A, Santos-Buitrago B, De Las Rivas J, Knapp M, Santos-García G, Talcott C. Epidermal Growth Factor Signaling towards Proliferation: Modeling and Logic Inference Using Forward and Backward Search. Biomed Res Int. 2017;2017:1809513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 32. | Santos-García G, Talcott CL, De Las Rivas J. Analysis of cellular proliferation and survival signaling by using two ligand/receptor systems modeled by Pathway Logic. In: Abate A, Safránek D, editors. Revised Selected Papers, volume 9271 of Lecture Notes in Computer Science. Hybrid Systems Biology - Fourth International Workshop, HSB 2015; 2015 Sep 4-5; Madrid, Spain. Springer, 2015: 226-245. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 33. | Knapp M, Briesemeister L, Eker S, Lincoln P, Poggio A, Talcott CL, Laderoute K. Pathway Logic helping biologists understand and organize pathway information. In: Markstein P, Xu Y, editors. Proceedings of the Fourth International IEEE Computer Society Computational Systems Bioinformatics Conference Workshops & Poster Abstracts (CSB 2005 Workshops); 2005 Aug 8-11; Stanford, United States. IEEE Computer Society, 2005: 155-156. [DOI] [Full Text] |
| 34. | Talcott CL, Dill DL. Multiple representations of biological processes. In: Corrado Priami C, Plotkin GD, editors. Transactions on Computational Systems Biology VI, volume 4220 of Lecture Notes in Computer Science. Springer, 2006: 221-245. [RCA] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 35. | Tenazinha N, Vinga S. A survey on methods for modeling and analyzing integrated biological networks. IEEE/ACM Trans Comput Biol Bioinform. 2011;8:943-958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 36. | Chylek LA, Harris LA, Faeder JR, Hlavacek WS. Modeling for (physical) biologists: an introduction to the rule-based approach. Phys Biol. 2015;12:045007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 37. | Chylek LA, Harris LA, Tung CS, Faeder JR, Lopez CF, Hlavacek WS. Rule-based modeling: a computational approach for studying biomolecular site dynamics in cell signaling systems. Wiley Interdiscip Rev Syst Biol Med. 2014;6:13-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 38. | Chylek LA, Stites EC, Posner RG, Hlavacek WS. Innovations of the rule-based modeling approach. In: Prokop A, Csukás B, editors. Systems Biology, Integrative Biology and Simulation Tools. Dordrecht: Springer, 2013: 273-300. [RCA] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | Harmer R, Le Cornec YS, Legare S, Oshurko E. Bio-Curation for Cellular Signalling: The KAMI Project. IEEE/ACM Trans Comput Biol Bioinform. 2019;16:1562-1573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 40. | Harris LA, Hogg JS, Tapia JJ, Sekar JA, Gupta S, Korsunsky I, Arora A, Barua D, Sheehan RP, Faeder JR. BioNetGen 2.2: advances in rule-based modeling. Bioinformatics. 2016;32:3366-3368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 135] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 41. | Hogg JS. Advances in rule-based modeling: Compartments, energy, and hybrid simulation, with application to sepsis and cell signaling. PhD thesis, University of Pittsburgh. 2013. Available from: http://d- scholarship.pitt.edu/id/eprint/19621. |
| 42. | Schaff JC, Vasilescu D, Moraru II, Loew LM, Blinov ML. Rule-based modeling with Virtual Cell. Bioinformatics. 2016;32:2880-2882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 43. | Smith AM, Xu W, Sun Y, Faeder JR, Marai GE. RuleBender: integrated modeling, simulation and visualization for rule-based intracellular biochemistry. BMC Bioinformatics. 2012;13 Suppl 8:S3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 44. | Khan S, Carvajal RD. Novel Approaches to the Systemic Management of Uveal Melanoma. Curr Oncol Rep. 2020;22:104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 45. | Gandini S, Sera F, Cattaruzza MS, Pasquini P, Picconi O, Boyle P, Melchi CF. Meta-analysis of risk factors for cutaneous melanoma: II. Sun exposure. Eur J Cancer. 2005;41:45-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 796] [Cited by in RCA: 831] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 46. | Mallet JD, Gendron SP, Drigeard Desgarnier MC, Rochette PJ. Implication of ultraviolet light in the etiology of uveal melanoma: A review. Photochem Photobiol. 2014;90:15-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 47. | Schadendorf D, van Akkooi ACJ, Berking C, Griewank KG, Gutzmer R, Hauschild A, Stang A, Roesch A, Ugurel S. Melanoma. Lancet. 2018;392:971-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 856] [Cited by in RCA: 968] [Article Influence: 138.3] [Reference Citation Analysis (0)] |
| 48. | Singh AD, Bergman L, Seregard S. Uveal melanoma: epidemiologic aspects. Ophthalmol Clin North Am. 2005;18:75-84, viii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 206] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 49. | Johansson PA, Brooks K, Newell F, Palmer JM, Wilmott JS, Pritchard AL, Broit N, Wood S, Carlino MS, Leonard C, Koufariotis LT, Nathan V, Beasley AB, Howlie M, Dawson R, Rizos H, Schmidt CW, Long GV, Hamilton H, Kiilgaard JF, Isaacs T, Gray ES, Rolfe OJ, Park JJ, Stark A, Mann GJ, Scolyer RA, Pearson JV, van Baren N, Waddell N, Wadt KW, McGrath LA, Warrier SK, Glasson W, Hayward NK. Whole genome landscapes of uveal melanoma show an ultraviolet radiation signature in iris tumours. Nat Commun. 2020;11:2408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 50. | De Fabo EC, Noonan FP, Fears T, Merlino G. Ultraviolet B but not ultraviolet A radiation initiates melanoma. Cancer Res. 2004;64:6372-6376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 179] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 51. | Shain AH, Bagger MM, Yu R, Chang D, Liu S, Vemula S, Weier JF, Wadt K, Heegaard S, Bastian BC, Kiilgaard JF. The genetic evolution of metastatic uveal melanoma. Nat Genet. 2019;51:1123-1130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 149] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 52. | Lo JA, Fisher DE. The melanoma revolution: from UV carcinogenesis to a new era in therapeutics. Science. 2014;346:945-949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 318] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 53. | Pho L, Grossman D, Leachman SA. Melanoma genetics: a review of genetic factors and clinical phenotypes in familial melanoma. Curr Opin Oncol. 2006;18:173-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 54. | Greene MH, Clark WH Jr, Tucker MA, Kraemer KH, Elder DE, Fraser MC. High risk of malignant melanoma in melanoma-prone families with dysplastic nevi. Ann Intern Med. 1985;102:458-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 288] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 55. | Rouse WB. Understanding the complexity of health. Syst Res Behav Sci. 2020;. [DOI] [Full Text] |
| 56. | Singh AD, Turell ME, Topham AK. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118:1881-1885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 941] [Cited by in RCA: 866] [Article Influence: 61.9] [Reference Citation Analysis (0)] |
| 57. | Damato B. Detection of uveal melanoma by optometrists in the United Kingdom. Ophthalmic Physiol Opt. 2001;21:268-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 58. | Nathan P, Cohen V, Coupland S, Curtis K, Damato B, Evans J, Fenwick S, Kirkpatrick L, Li O, Marshall E, McGuirk K, Ottensmeier C, Pearce N, Salvi S, Stedman B, Szlosarek P, Turnbull N; United Kingdom Uveal Melanoma Guideline Development Working Group. Uveal Melanoma UK National Guidelines. Eur J Cancer. 2015;51:2404-2412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 83] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 59. | Dinarès Fernández CM. Estudio de la vía de señalización de 4E-BP1. Implicaciones pronósticas y terapéuticas en el melanoma uveal. PhD thesis, Autonomous University of Barcelona. 2017. Available from: https://hdl.handle.net/10803/405446. |
| 60. | Aughton K, Shahidipour H, Djirackor L, Coupland SE, Kalirai H. Characterization of Uveal Melanoma Cell Lines and Primary Tumor Samples in 3D Culture. Transl Vis Sci Technol. 2020;9:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 61. | Weis E, Salopek TG, McKinnon JG, Larocque MP, Temple-Oberle C, Cheng T, McWhae J, Sloboda R, Shea-Budgell M. Management of uveal melanoma: a consensus-based provincial clinical practice guideline. Curr Oncol. 2016;23:e57-e64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 62. | Fernandes BF, Belfort RN, Di Cesare S, Burnier MN Jr. Circulating uveal melanoma cells: should we test for them? Can J Ophthalmol. 2008;43:155-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 63. | Li Y, Shi J, Yang J, Ge S, Zhang J, Jia R, Fan X. Uveal melanoma: progress in molecular biology and therapeutics. Ther Adv Med Oncol. 2020;12:1758835920965852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 64. | Blasi MA, Laguardia M, Tagliaferri L, Scupola A, Villano A, Caputo CG, Pagliara MM. Brachytherapy Alone or With Neoadjuvant Photodynamic Therapy for Amelanotic Choroidal Melanoma: Functional Outcomes and Local Tumor Control. Retina. 2016;36:2205-2212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 65. | Seibel I, Cordini D, Rehak M, Hager A, Riechardt AI, Böker A, Heufelder J, Weber A, Gollrad J, Besserer A, Joussen AM. Local Recurrence After Primary Proton Beam Therapy in Uveal Melanoma: Risk Factors, Retreatment Approaches, and Outcome. Am J Ophthalmol. 2015;160:628-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 66. | Harbour JW, Char DH, Kroll S, Quivey JM, Castro J. Metastatic risk for distinct patterns of postirradiation local recurrence of posterior uveal melanoma. Ophthalmology. 1997;104:1785-92; discussion 1792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 55] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 67. | Singh AD, Shields CL, Shields JA. Prognostic factors in uveal melanoma. Melanoma Res. 2001;11:255-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 112] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 68. | Bell DJ, Wilson MW. Choroidal melanoma: natural history and management options. Cancer Control. 2004;11:296-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 69. | Durante MA, Rodriguez DA, Kurtenbach S, Kuznetsov JN, Sanchez MI, Decatur CL, Snyder H, Feun LG, Livingstone AS, Harbour JW. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat Commun. 2020;11:496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 298] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 70. | Helgadottir H, Höiom V. The genetics of uveal melanoma: current insights. Appl Clin Genet. 2016;9:147-155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 71. | Moore AR, Ran L, Guan Y, Sher JJ, Hitchman TD, Zhang JQ, Hwang C, Walzak EG, Shoushtari AN, Monette S, Murali R, Wiesner T, Griewank KG, Chi P, Chen Y. GNA11 Q209L Mouse Model Reveals RasGRP3 as an Essential Signaling Node in Uveal Melanoma. Cell Rep. 2018;22:2455-2468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 72. | Amirouchene-Angelozzi N, Schoumacher M, Stern MH, Cassoux N, Desjardins L, Piperno-Neumann S, Lantz O, Roman-Roman S. Upcoming translational challenges for uveal melanoma. Br J Cancer. 2015;113:1249-1253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 73. | Li J, Liu X, Li C, Wang W. miR-224-5p inhibits proliferation, migration, and invasion by targeting PIK3R3/AKT3 in uveal melanoma. J Cell Biochem. 2019;120:12412-12421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 74. | Talcott CL. Formal executable models of cell signaling primitives. In: Margaria T, Steffen B, editors. Proceedings of the Leveraging Applications of Formal Methods Second International Symposium, ISoLA 2006; 2006 Nov 15-19; Paphos, Cyprus. IEEE, 2006: 298-302. [DOI] [Full Text] |
| 75. | Talcott C, Eker S, Knapp M, Lincoln P, Laderoute K. Pathway logic modeling of protein functional domains in signal transduction. Pac Symp Biocomput. 2004: 568-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 76. | Santos-García G, Talcott CL, Riesco A, Santos-Buitrago B, De Las Rivas J. Role of nerve growth factor signaling in cancer cell proliferation and survival using a reachability analysis approach. In: Mohamad MS, Rocha MP, Fdez-Riverola F, Mayo FJD, De Paz JF, editors. Volume 477 of Advances in Intelligent Systems and Computing. 10th International Conference on Practical Applications of Computational Biology & Bioinformatics, PACBB 2016; 2016 Jun 1-3; Sevilla, Spain. Springer, 2016: 173-181. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 77. | Talcott CL. Symbolic modeling of signal transduction in Pathway Logic. In: Perrone LF, Lawson B, Liu J, Wieland FP, editors. Proceedings of the Winter Simulation Conference, WSC 2006; 2006 Dec 3-6; Monterey, United States. WSC, 2006: 1656-1665. [DOI] [Full Text] |
| 78. | Meseguer J. Conditional rewriting logic as a unified model of concurrency. Theor Comput Sci. 1992;96:73–155. [DOI] [Full Text] |
| 79. | Santos-García G, Palomino M, Verdejo A. Rewriting logic using strategies for neural networks: An implementation in Maude. In: Corchado JM, Rodríguez S, Llinas J, Molina JM, editors. Volume 50 of Advances in Soft Computing. Proceedings of the International Symposium on Distributed Computing and Artificial Intelligence, DCAI 2008; 2008 Oct 22-24; Salamanca, Spain. Springer, 2009: 424-433. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 80. | Clavel M, Durán F, Eker S, Lincoln P, Martí-Oliet N, Meseguer J, Talcott CL. All about Maude - A high-performance logical framework, how to specify, program and verify systems in Rewriting Logic, volume 4350 of Lecture Notes in Computer Science. Springer, 2007. [DOI] [Full Text] |
| 81. | Riesco A, Santos-Buitrago B, Knapp M, Santos-García G, Galilea EH, Talcott CL. Fuzzy matching for cellular signaling networks in a choroidal melanoma model. In: Panuccio G, Rocha M, Fdez-Riverola F, Mohamad MS, Casado-Vara R, editors. Volume 1240 of Advances in Intelligent Systems and Computing. Practical Applications of Computational Biology & Bioinformatics, 14th International Conference (PACBB 2020); 2020 Jun 17-19; L’Aquila, Italy. Springer, 2020: 80-90. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 82. | Martí-Oliet N, Ölveczky PC, Talcott CL. Martí-Oliet N, Ölveczky PC, Talcott CL. Logic, Rewriting, and Concurrency - Essays dedicated to José Meseguer on the occasion of his 65th birthday, volume 9200 of Lecture Notes in Computer Science. Springer, 2015. [DOI] [Full Text] |
| 83. | Santos-Buitrago B, Riesco A, Knapp M, Alcantud JCR, Santos-García G, Talcott C. Soft Set Theory for Decision Making in Computational Biology under Incomplete Information. IEEE Access. 2019;7:18183-18193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 84. | Santos-Buitrago B, Galilea EH. Signaling transduction networks in choroidal melanoma: A symbolic model approach. In: Fdez-Riverola F, Rocha M, Mohamad MS, Zaki N, Castellanos-Garzón JA, editors. Volume 1005 of Advances in Intelligent Systems and Computing. Practical Applications of Computational Biology and Bioinformatics, 13th International Conference, PABCC 2019; 2019 Jun 26-28; Ávila, Spain. Springer, 2019: 96-104. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 85. | Krantz BA, Dave N, Komatsubara KM, Marr BP, Carvajal RD. Uveal melanoma: epidemiology, etiology, and treatment of primary disease. Clin Ophthalmol. 2017;11:279-289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 248] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 86. | Patel M, Smyth E, Chapman PB, Wolchok JD, Schwartz GK, Abramson DH, Carvajal RD. Therapeutic implications of the emerging molecular biology of uveal melanoma. Clin Cancer Res. 2011;17:2087-2100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 87. | Xu X, Zong Y, Gao Y, Sun X, Zhao H, Luo W, Jia S. VEGF Induce Vasculogenic Mimicry of Choroidal Melanoma through the PI3k Signal Pathway. Biomed Res Int. 2019;2019:3909102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |