Published online Aug 28, 2020. doi: 10.13105/wjma.v8.i4.309
Peer-review started: June 8, 2020
First decision: July 3, 2020
Revised: July 17, 2020
Accepted: August 27, 2020
Article in press: August 27, 2020
Published online: August 28, 2020
Processing time: 93 Days and 11.3 Hours
Meta-analysis, a form of quantitative review, is an attempt to combine data from multiple independent studies to improve statistical power. Because of the complexity of process involved in study selection, data analysis, and evaluation of bias and heterogeneity, checklists have been prepared by the Institutes of Medicine (IOM), Preferred Reporting Items for Systemic Reviews and Meta-analyses (PRISMA), and Meta-analyses of Observational Studies in Epidemiology (MOOSE) to standardize the reporting quality of a meta-analysis.
To use these checklists to assess the reporting quality of the coronavirus disease-2019 (COVID-19) meta-analysis literature relevant to laboratory hematology.
After a search of the literature 19 studies were selected for analysis, including 10 studies appearing in the preprint literature (studies that can be identified by database search but have not yet completed peer review).
The average IOM (76% of required elements completed), PRISMA (75% of required elements completed), and MOOSE (60% of required elements completed) scores enumerated demonstrated a reporting quality inferior to that of earlier reports of pathology and medicine meta-analyses. There was no statistically significant difference in performance between accepted/ published and preprint studies. Comparison of the results of PRISMA and MOOSE studies demonstrated a weak positive correlation (Pearson’s correlation coefficient = 0.39).
The most common deficits in the studies included sensitivity analysis, assessment for bias, and details of the search strategy. Although the COVID-19 laboratory hematology meta-analysis literature can be a helpful source of information, readers should be aware of these reporting quality deficits.
Core Tip: The Institutes of Medicine, Preferred Reporting Items for Systemic Reviews and Meta-analyses, and Meta-analyses of Observational Studies in Epidemiology checklists were created to standardize the reporting quality of a meta-analysis. The purpose of this study was to use these checklists to assess the reporting quality of the coronavirus disease-2019 meta-analysis literature relevant to laboratory hematology.
- Citation: Frater JL. Importance of reporting quality: An assessment of the COVID-19 meta-analysis laboratory hematology literature. World J Meta-Anal 2020; 8(4): 309-319
- URL: https://www.wjgnet.com/2308-3840/full/v8/i4/309.htm
- DOI: https://dx.doi.org/10.13105/wjma.v8.i4.309
Meta-analysis, the examination of data from multiple independent studies of the same subject, is a useful form of quantitative review that can provide improved statistical power compared to studies with smaller numbers of subjects and demonstrate the presence or lack of consensus regarding a specific scientific question[1]. In recent years, the number of published meta-analyses has increased, particularly in the realm of clinical medicine, and they have become important sources of information for practitioners, especially in areas where information is rapidly evolving.
In pathology and laboratory medicine, meta-analyses are published less frequently compared to other areas of clinical medicine. Kinzler and Zhang, in their survey of the meta-analysis literature in pathology journals compared to medicine journals, note a significantly larger percentage of publication space dedicated to meta-analyses in medicine journals[1]. This is despite the proven high quality of meta-analyses in both journal categories, as evidenced by similar adjusted citation ratios (which they defined as article’s citation count divided by the mean citations for the meta-analysis, review, and original research articles published in the same journal the same year)[1].
Because meta-analyses are an important source of information for clinicians and others, it is essential that they are formatted to easily allow the reader to assess their strengths and weaknesses. Several checklists have been established by national and international committees, including the Institutes of Medicine (IOM), Preferred Reporting Items for Systemic Reviews and Meta-analyses (PRISMA), and Meta-analyses of Observational Studies in Epidemiology (MOOSE)[2-4]. A recent survey by Liu et al[5] using the PRISMA criteria noted that the reporting quality for a sampling of medicine meta-analyses was higher than that of pathology meta-analyses. The overall reporting quality for laboratory hematology-focused meta-analyses was not specifically addressed[5].
The coronavirus disease-2019 (COVID-19) pandemic, which originated in the city of Wuhan in the Hubei Province of China in December 2019 quickly spread to Europe and then to North America[6,7]. In an effort to study the disease and improve the world health community’s response, over 30000 papers have been added to the medical literature since December 2019, based on a search of the PubMed database for the keyword “COVID-19” conducted on July 16, 2020. In a situation such as this, it is essential for the practicing clinician to have access to reliable studies with good statistical power, hence the need for meta-analyses with high reporting quality. Laboratory hematology is an essential component of the medical response to COVID-19 since several biomarkers of infection derived from the complete blood count (CBC) and coagulation testing are of proven utility in assessing prognosis and likely outcome[8-10]. As in all quickly evolving fields, a large fraction of the accessible medical COVID medical literature appears in the form of preprint publications. These are manuscripts that are indexed in services such as Google Scholar, but have not yet completed the peer-review process. The purpose of this study is two-fold; to assess the reporting quality of COVID-19 meta-analyses focused on laboratory hematology and to compare the reporting quality of published studies of COVID-19 to the preprint literature.
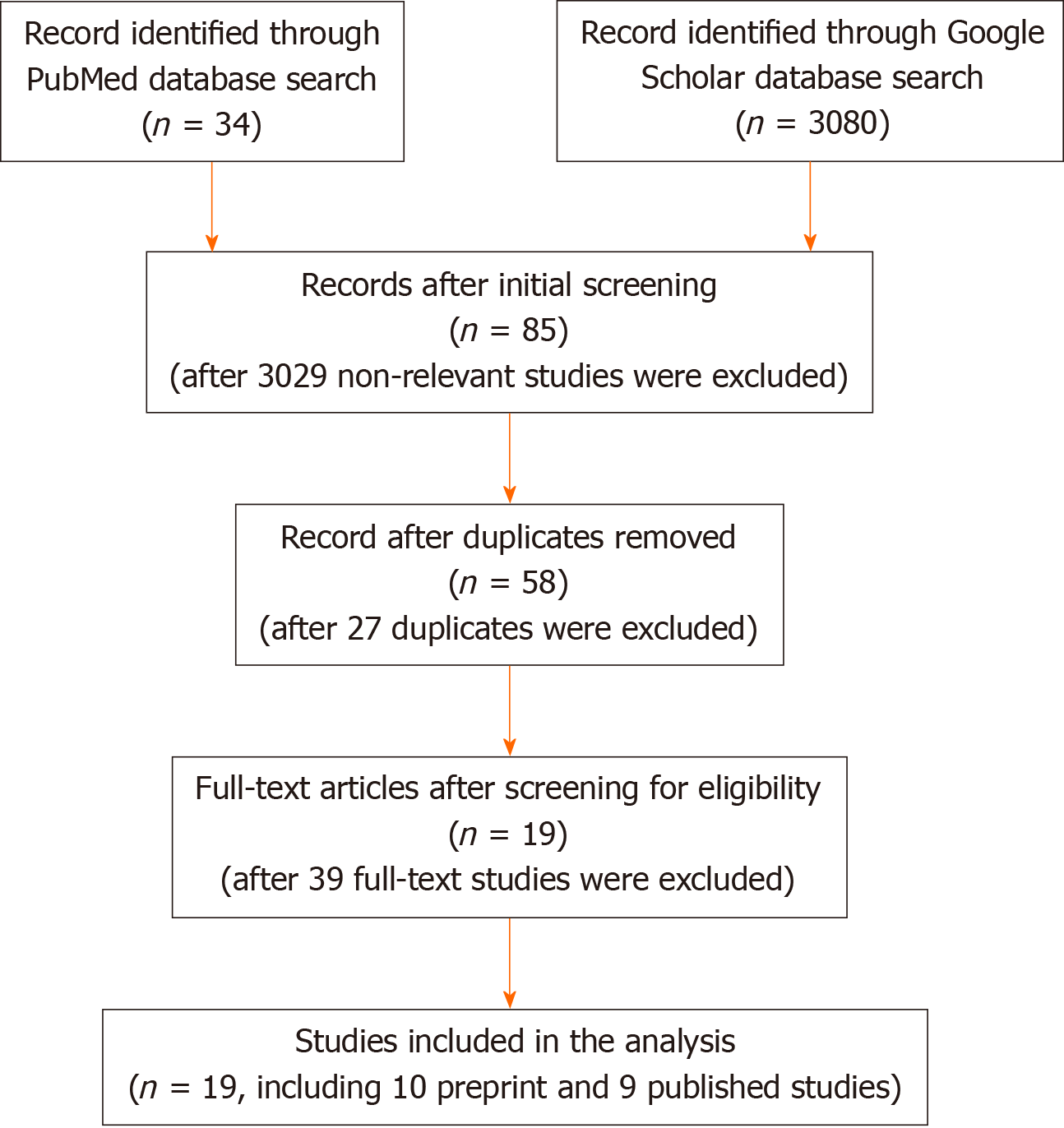
The study selection processes is summarized in Figure 1. A search was conducted in PubMed and Google Scholar using the search terms “COVID-19” OR “COVID”, “SARS-CoV-2”, OR “coronavirus” AND “meta-analysis”, which yielded 34 entries in PubMed and 3080 in Google Scholar (total = 3114 studies). Initial screening for letters to the editor, editorials, and non-meta-analysis reviews removed 3029 publications, with 85 entries remaining for further consideration. After removal of 27 duplicate entries, 58 publications remained. The full text of the remaining 58 studies were examined for content, and 39 studies that fell out of scope for further consideration were removed, leaving 19 studies for the analysis.
The studies were separated into published studies (n = 9, Table 1)[11-19] and manuscripts appearing in the preprint literature (n = 10, Table 1)[20-28]. For the purposes of this study, preprint literature refers to manuscripts discoverable in the Google Scholar database which have been submitted for publication and are assigned an identifier through a service such as doi.org or preprints.org but have not completed the peer-review process.
| Ref. | Country1 | No. of patients | Evaluated hematologic parameters |
| Published studies | |||
| Borges et al[11] | Multinational, predominantly China | 59254 | WBC, ANC, ALC, PLT, D-Dimer |
| Cao et al[12] | China | 46959 | WBC, ALC |
| Fu et al[13] | Not stated, likely all China | 3600 | WBC, ALC, PLT, D-dimer |
| Henry et al[14] | China, Singapore | 2984 | WBC, ANC, ALC, MONO, EOS, HGB, PT, PTT, D-dimer |
| Lagunes-Rangel[15] | China | 828 | Estimate of N/L ratio |
| Li et al[16] | China | 1995 | WBC |
| Lippi et al[17] | China, Singapore | 1099 | PLT |
| Rodriguez-Morales et al[18] | China, Australia | 2874 | WBC, ALC, HGB |
| Zhu et al[19] | China | 3062 | WBC, ALC, D-dimer |
| Preprint studies | |||
| Arabi et al[20] | China | 50 | WBC |
| Ebrahami et al[21] | China | 2217 | WBC, ANC, ALC, HGB, PLT, PT, PTT, D-Dimer |
| Han et al[22] | China | 1208 | ALC, ANC, PLT, PT, PTT, D-Dimer |
| Heydari et al[23] | China, S. Korea | 49504 | WBC, ANC, ALC, D-dimer |
| Ma et al[24] | China | 53000 | ALC, PLT, D-dimer |
| Nasiri et al[25] | China, Germany | 4679 | WBC, ANC, ALC, HGB, PLT |
| Pormohammad et al[26] | China | 52251 | WBC, ALC, ANC, PLT, HGB |
| Xu et al[27] | China | 4062 | WBC, ANC,ALC, PLT, D-dimer |
| Zhang et al[28] | Not stated, likely all China | 275 | WBC, ALC |
The studies were then evaluated using the IOM, PRISMA, and MOOSE criteria. The IOM has compiled a list of 5 required elements that serve as recommended standards for meta-analysis (Table 2)[2]. The PRISMA group compiled a list of 27 checklist items to facilitate the assessment of the reporting quality of meta-analyses[3]. The MOOSE criteria consist of a 34-point checklist categorized under 5 divisions[4]. The criteria were evaluated for each study, and a numeric score was assigned based on the sum total of positive results for each element of the IOM, PRISMA and MOOSE checklists.
| Required element | Papers meeting this standard (total number and percentage) | Published/ accepted papers meeting this standard (total number and percentage) | Preprint papers meeting this standard (total number and percentage) |
| Explain why a pooled estimate might be useful to decision makers | 9/19 (47%) | 5/9 (56%) | 4/10 (40%) |
| Use expert methodologists to develop, execute, and peer review the meta-analyses | 15/19 (79%) | 7/9 (78%) | 8/10 (80%) |
| Address heterogeneity among study effects | 18/19 (95%) | 8/9 (89%) | 10/10 (100%) |
| Accompany all estimates with measures of statistical uncertainty | 19/19 (100%) | 9/9 (100%) | 10/10 (100%) |
| Assess the sensitivity of conclusions to changes in the protocol, assumptions, and study selection (sensitivity analysis) | 12/19 (63%) | 5/9 (56%) | 7/10 (70%) |
The mean PRISMA and MOOSE scores for the accepted/published and preprint studies were compared using the student 2-tail t-test, with significance defined as P < 0.05. The PRISMA and MOOSE scores were compared using Pearson’s correlation coefficient. All statistics were calculated using Excel (Microsoft, Redmond, WA, United States).
Qualitative features of the studies are summarized in Table 1. Most cases (17 of 19, 89%) were from Chinese patient populations. For the remaining 2 studies, the national origin of the patient populations was not defined, but given the affiliations of the authors, the patient cohorts were also likely from China. The number of patients in each study was highly variable, ranging from 50 to 59254. The hematology data reported in the studies was heterogeneous. The most common evaluated tests were white blood cell count (15 studies), absolute lymphocyte count (15 studies), and platelet count (10 studies).
Because of the limited number of reporting elements in the IOM checklist (Table 2), a comparison with the PRISMA (Table 3) and MOOSE (Table 4) checklists was not performed. The mean IOM score was 3.8/5 (76%) for all studies. The average scores for preprint (4.0/5, 80%) and accepted/ published (3.5, 70%) studies was similar, and there was no statistically significant difference between the two groups (P > 0.05). Reviewing the IOM required elements, the most common deficiencies were in explaining why a pooled estimate might be useful to decision makers and lack of sensitivity analysis.
| Item number | Element | Papers meeting this standard (total number and percentage) | Published/ accepted papers meeting this standard (total number and percentage) | Preprint papers meeting this standard (total number and percentage) |
| 1 | Title | 19/19 (100%) | 8/9 (89%) | 10/10 (100%) |
| 2 | Structured summary | 18/19 (95%) | 8/9 (89%) | 10/10 (100%) |
| Introduction | ||||
| 3 | Rationale | 16/19 (84%) | 8/9 (80%) | 8/10 (89%) |
| 4 | Objectives | 17/19 (89%) | 9/9 (90%) | 8/10 (89%) |
| Methods | ||||
| 5 | Protocol/Registration | 16/19 (84%) | 8/9 (89%) | 8/10 (78%) |
| 6 | Eligibility criteria | 17/19 (89%) | 8/9 (89%) | 9/10 (89%) |
| 7 | Information sources | 18/19 (95%) | 9/9 (100%) | 9/10 (89%) |
| 8 | Search | 18/19 (95%) | 9/9 (100%) | 9/10 (89%) |
| 9 | Study selection | 19/19 (100%) | 9/9 (100%) | 10/10 (100%) |
| 10 | Data collection process | 18/19 (95%) | 8/9 (89%) | 10/10 (100%) |
| 11 | Data items | 17/19 (89%) | 8/9 (89%) | 9/10 (89%) |
| 12 | Risk of bias in individual studies | 10/19 (53%) | 4/9 (44%) | 6/10 (56%) |
| 13 | Summary measures | 15/19 (79%) | 6/9 (66%) | 8/10 (78%) |
| 14 | Synthesis of results | 16/19 (84%) | 7/9 (78%) | 9/10 (89%) |
| 15 | Risk of bias across studies | 2/19 (11%) | 2/9 (22%) | 0/10 (0) |
| 16 | Additional analyses | 2/19 (11%) | 1/9 (11%) | 1/10 (10%) |
| Results | ||||
| 17 | Study selection | 19/19 (100%) | 9/9 (100%) | 10/10 (100%) |
| 18 | Study characteristics | 19/19 (100%) | 9/9 (100%) | 10/10 (100%) |
| 19 | Risk of bias within studies | 12/19 (63%) | 5/9 (56%) | 7/10 (70%) |
| 20 | Results of individual studies | 11/19 (58%) | 5/9 (56%) | 6/10 (60%) |
| 21 | Synthesis of results | 16/19 (84%) | 8/9 (89%) | 8/10 (80%) |
| 22 | Risk of bias across studies | 9/19 (47%) | 5/9 (56%) | 4/10 (40%) |
| 23 | Additional analysis | 0/19 (0) | 0/9 (0) | 0/10 (0) |
| Discussion | ||||
| 24 | Summary of evidence | 19/19 (100%) | 9/9 (100%) | 10/10 (100%) |
| 25 | Limitations | 16/19 (84%) | 7/9 (78%) | 9/10 (90%) |
| 26 | Conclusions | 19/19 (100%) | 9/9(100%) | 10/10 (100%) |
| Funding | ||||
| 27 | Funding | 7/19 (37%) | 3/9 (33%) | 4/10 (40%) |
| Checklist | Number | Papers meeting this standard (total number and percentage) | Published/ accepted papers meeting this standard (total number and percentage) | Preprint papers meeting this standard (total number and percentage) |
| I. Reporting of background | ||||
| A. Problem definition | 10/19 (53%) | 7/9 (78%) | 3/10 (30%) | |
| B. Hypothesis statement | 2/19 (11%) | 1/9 (11%) | 1/10 (10%) | |
| C. Description of study outcome(s) | 19/19 (100%) | 10/9 (100%) | 9/10 (90%) | |
| D. Type of exposure or intervention used | 18/19 (95%) | 9/9 (100%) | 8/10 (80%) | |
| E. Type of study designs used | 18/19 (95%) | 9/9 (100%) | 9/10 (90%) | |
| F. Study population | 18/19 (95%) | 9/9 (100%) | 9/10 (90%) | |
| II. Reporting of search strategy | ||||
| A. Qualifications of searchers | 0/19 (0) | 0/9 (0) | 0/10 (0) | |
| B. Search strategy | 17/19 (89%) | 9/9 (100%) | 8/10 (80%) | |
| C. Effort to include all available studies | 10/19 (53%) | 7/9 (78%) | 3/10 (30%) | |
| D. Databases and registries searched | 17/19 (89%) | 7/9 (78%) | 10/10 (100%) | |
| E. Search software used | 8/19 (42%) | 4/9 (44%) | 4/10 (40%) | |
| F. Use of hand searching | 2/19 (11%) | 1/9 (11%) | 1/10 (10%) | |
| G. List of citations located and those excluded | 10/19 (53%) | 5/9 (56%) | 5/10 (50%) | |
| H. Method of addressing articles published in languages other than English | 0/19 (0) | 0/9 (0) | 0/10 (0) | |
| I. Method of handling abstracts and unpublished studies | 0/19 (0) | 0/9 (0) | 0/10 (0) | |
| J. Description of any contact with authors | 0/19 (0) | 0/9 (0) | 0/10 (0) | |
| III. Reporting of methods | ||||
| A. Description of relevance or appropriateness of studies assembled for assessing the hypothesis to be tested | 8/19 (42%) | 4/9 (44%) | 4/10 (40%) | |
| B. Rationale for the selection and coding of data | 13/19 (68%) | 7/9 (78%) | 6/10 (60%) | |
| C. Documentation of how data were classified and coded | 12/19 (63%) | 8/9 (89%) | 4/10 (40%) | |
| D. Assessment of confounding | 1/19 (5%) | 0/9 (0) | 1/10 (10%) | |
| E. Assessment of study quality | 16/19 (84%) | 7/9 (78%) | 9/10 (90%) | |
| F. Assessment of heterogeneity | 18/19 (95%) | 8/9 (89%) | 10/10 (100%) | |
| G. Description of statistical methods | 19/19 (100%) | 9/9 (100%) | 10/10 (100%) | |
| H. Provision of appropriate tables and graphics | 18/19 (95%) | 9/9 (100%) | 9/10 (90%) | |
| IV. Reporting of results | ||||
| A. Graphic summarizing individual study estimates and overall estimate | 19/19 (100%) | 9/9 (100%) | 10/10 (100%) | |
| B. Table giving descriptive information for each study included | 16/19 (84%) | 7/9 (78%) | 9/10 (90%) | |
| C. Results of sensitivity testing (e.g, subgroup analysis) | 12/19 (63%) | 7/9 (78%) | 5/10 (50%) | |
| D. Indication of statistical uncertainty of findings | 17/19 (89%) | 8/9 (89%) | 9/10 (90%) | |
| E. Reporting of discussion should include | ||||
| 1. Quantitative assessment of bias (e.g, publication bias) | 11/19 (58%) | 4/9 (44%) | 7/10 (70%) | |
| 2. Justification for exclusion (eg, exclusion of non–English-language citations) | 3/19 (16%) | 1/9 (11%) | 2/10 (20%) | |
| 3. Assessment of quality of included studies | 12/19 (63%) | 4/9 (44%) | 8/10 (80%) | |
| V. Reporting of conclusions | ||||
| A. Consideration of alternative explanations for observed results | 1/19 (11%) | 0/9 (0) | 1/10 (10%) | |
| B. Generalization of the conclusions (i.e, appropriate for the data presented and within the domain of the literature review) | 19/19 (100%) | 9/9 (100%) | 10/10 (100%) | |
| C. Guidelines for future research | 8/19 (42%) | 6/9 (66%) | 2/10 (20%) | |
Due to the larger number of reporting elements in the PRISMA and MOOSE checklists a more robust comparison could be performed. The average PRISMA score for all studies was 20.3/27 (75%) (median = 22/27, 81%).The average scores of the accepted/published (mean = 20.4/27, 76% median = 21.5/27, 80%) and preprint (mean = 20.2/27, 75%, median = 22/27, 81%) groups were similar (student t-test, P > 0.05). The most common elements which were lacking were checklist numbers 15 (methods: risk of bias across studies), 16 (methods: additional analyses), 22 (results: risk of bias across studies), and 23 (results: risk of bias across studies). The average MOOSE scores for all studies was 19.9/34, 60% (median = 20/34, 60%).The average scores of the accepted/published (mean = 20.6, 61% median = 21/34, 62%) and preprint (mean = 19.1, 56% median = 19, 56%) groups were similar (student t-test, P > 0.05). The most common elements which were lacking were II.A [Qualifications of searchers (e.g., librarians and investigators)], II.H (Method of addressing articles published in languages other than English, II.I (Method of handling abstracts and unpublished studies) and II.J (Description of any contact with authors).
To determine the degree to which the PRISMA and MOOSE scores correlated, analysis using Pearson’s correlation coefficient was performed. The resulting coefficient, 0.39, suggests a weak positive correlation.
Narrative, nonquantitative review papers have existed in the medical literature for many years and are an important source for succinctly reported and up-to-date information for clinicians and others interested in patient care and other issues. In recognition of the importance of the evidence-based approach to the dissemination of medical information, authors added increasingly rigorous approaches to their publications to provide quantitative information, minimize bias, identify knowledge gaps in the regarding a subject, and provide guidance for further growth of the area of study. This trend resulted in the development of the meta-analysis[29].
Meta-analysis is a modification and attempted improvement of more traditional forms of review publication Meta-analysis attempts to move beyond the narrative review process by adding numeric data synthesized from previously published data[30]. By combining data from more than one study, there is an obvious improvement in statistical power. Meta-analysis has been widely employed in the behavioral science and clinical medicine literatures but has been underutilized in the pathology and laboratory medicine literature. Kinzler and Zhang published a study comparing the use of meta-analysis in the diagnostic pathology literature compared to the clinical medicine literature and noted that meta-analyses comprised < 1% of diagnostic pathology articles compared to 4%-6% of the clinical medicine literature[1]. Despite their relatively low numbers, meta-analyses in the diagnostic pathology literature were highly cited, with a citation rate similar to that of meta-analyses appearing in the clinical medicine literature[1]. This finding is also noted in the current study: although numerous studies have been published addressing the laboratory hematologic aspects of COVID-19, the number of meta-analyses is low and comprises < 1% of the published literature in this area.
To be successful, the meta-analysis must address several elements[29]: (1) The question must be stated unambiguously; (2) A search of the medical literature must be performed in a comprehensive way; (3) The articles identified by the search must be screened; (4) The appropriate data must be extracted from the selected papers; (5) An assessment of the quality of the information is performed, by a review of the contents of the manuscripts and the Grading of Recommendations Assessment, Development and Evaluation (GRADE) criteria[30]; (6) Determine whether the data in each publication are heterogeneous; (7) Determine summary effect size as odds ratio and generate graphical depictions of data, for example as a forest plot; (8) Assess for publication bias using funnel plot or some other mechanism; and (9) Conduct subset analysis to look for subsets of groups that capture the summary effect.
Because of the complexity of design and execution of meta-analyses, there are numerous opportunities to introduce biases and other errors that may significantly alter the outcome. To make the reporting of data and statistical analysis in meta-analyses transparent to the reader and to clearly advertise the limits of the data used in the study, 3 checklist systems have been promulgated to list the major elements that researchers should use to structure their work.
The first of these systems, the IOM checklist, was created by a committee by the United States Institutes of Medicine. This is a relatively simple 5-point checklist that broadly addresses the reporting of the planning and execution of meta-analyses[2]. The Institutes of Medicine, along with a large number of journals and other publishers, later endorsed the PRISMA statement, which addresses these issues in a more granular fashion[3]. Anther checklist, the MOOSE guidelines, may also be applied to evaluate reporting quality of systematic reviews including meta-analyses[4]. In the reported literature, PRISMA guidelines are utilized more frequently than MOOSE guidelines. In a survey of the medical literature by Fleming et al[31], the vast majority of publications used PRISMA guidelines, compared to MOOSE guidelines, which were cited in only 17% of reviews. Fleming et al[31] note that although there is a high degree of overlap between the MOOSE and PRISMA checklists, MOOSE provides more advice about features such as the search strategy and interpretation of the results of the review, both of which may introduce bias if not adequately addressed[31,32].
In the current study the most common deficiencies were (1) lack of an articulated rationale for why a pooled analysis is necessary; (2) lack of detail of how to address the use of data that has not been peer reviewed; (3) a lack of sensitivity analysis; and (4) a lack of assessment of studies for bias. Although the rationale for why a meta-analysis is performed is generally obvious (e.g., improved statistical power, identification of a consensus/lack of consensus regarding a specific clinical question) it is not explicitly articulated in a significant number of studies included in this survey. The lack of transparency about the use of non-English language literature and preprint and other non-peer reviewed materials may be problematic, in particular in COVID-19 studies. Sensitivity analysis is a fundamental element of meta-analysis and provides an estimate of the appropriateness of the assumptions made by the analysis[29]. Bias can be introduced into a study in many ways, most commonly by publication bias, in which the medical literature has an underrepresentation of studies with negative findings[29].
The overall reporting of quality in the pathology literature appears to lag behind that for clinical medicine[5]. Liu et al[5] compared the reporting quality of a group of diagnostic pathology meta-analyses to a group published in clinical medicine journals using the PRISMA checklist, and found a higher average PRISMA score for the medicine studies that was statistically significant (P < 0.01). The average PRISMA score for the COVID-19 meta-analyses in the current study (20.3/27, 75% of items addressed) is below that for both groups analyzed by Liu et al[5]. This reflects a significant weakness in the COVID-19 meta-analysis laboratory hematology literature, since the potential strengths of the meta-analysis approach as a force multiplier for evidence-based medicine requires good reporting quality[5].
It is important to note the assessment of reporting quality is not synonymous with assessment of methodological quality of a meta-analysis. The purpose of reporting quality guidelines is to provide an appropriate framework to the authors of meta-analyses and other systematic reviews so that their data and statistical analysis is reported in an unambiguous way. The assessment of methodological quality is a separate exercise and can only proceed if the data can be unambiguously extracted from the publication. The methodological assessment of systematic reviews is addressed by other guidelines such as QUADAS and QUADAS-2[33]. Due to the apparent suboptimal average reporting quality of COVID-19 laboratory hematology meta-analyses literature, the ability of the reader to assess methodological quality is limited in many cases.
In academic publishing, a preprint is the version of a manuscript that has been submitted for publication but has not yet finished the peer review process. In recent years, publishers and others have electronically posted preprint manuscripts to rapidly disseminate scientific knowledge. In addition, studies that have been uploaded to dedicated servers but not submitted for peer review are also included in the category of preprints. Preprints are particularly useful in fields such as COVID-19, which are rapidly evolving and are of intense clinical and scientific interest.
Since preprints are widely accessible, it would be important for readers to be aware of their quality compared to studies published in the peer review literature. Although it would be assumed that the reporting quality of the peer review process would be higher than the comparable preprint literature since the purpose of peer review is to permit scrutiny of one’s work by experts[34], there have apparently been no studies in the peer review literature that directly compare the reporting quality of clinical studies in the preprint and published literature. A single study in the preprint literature (Carneiro et al[35]) has attempted to address this question. The authors compared a sample of studies identified in the bioRXIV preprint server with studies identified in a Medline (PubMed interface) search. They also compared a group of preprint studies with their final versions. Carneiro et al[35] identified a small increase in quality in the published studies compared to the preprint group.
In the current study, using the PRISMA and MOOSE criteria, a significant difference was not identified comparing the preprint and published studies in the COVID-19 meta-analysis literature. Taken together, these findings suggest that the peer review process itself does not guarantee an improvement in quality, and authors should take the initiative to conform to reporting quality norms.
This study represents an attempt to assess the overall reporting quality of the laboratory hematology COVID-19 meta-analysis literature. Using the IOM, PRISMA, and MOOSE, guidelines, there were consistent deficits in the reporting of bias and sensitivity. The results for the preprint and published literature were similar and suggest that the preprint literature on this subject is not decidedly inferior to the published literature. Because of the suboptimal reporting quality, it is important for clinicians and others to carefully assess the individual studies used in a given meta-analysis for evidence of bias or other methodological flaws that have not been reported by the authors. Although there is a positive correlation between the PRISMA and MOOSE guidelines, it is relatively weak. This implies that authors of meta-analyses should consider using both systems to increase the strength of the reporting quality of their studies.
Meta-analyses, which are underutilized in pathology and laboratory medicine, combine the data from multiple studies to produce a publication with increased statistical power. It is important for readers of meta-analyses to have the information in these studies reported in a transparent fashion. Hence the Institutes of Medicine (IOM), Preferred Reporting Items for Systemic Reviews and Meta-analyses (PRISMA), and Meta-analyses of Observational Studies in Epidemiology (MOOSE) checklists have been promulgated to standardize the reporting of meta-analyses.
Several parameters evaluated by the hematology laboratory have been identified as potential biomarkers of prognosis and outcome in the coronavirus disease 2019 (COVID-19). The data from many of these studies have been pooled and published as meta-analyses. Many of these studies have been identified in the preprint literature (studies that have not yet completed peer review). The reporting quality of this body of work is unknown.
The purposes of this study were to 1) evaluate the reporting quality of laboratory hematology-focused COVID-19 meta-analyses using the IOM, PRISMA, and MOOSE checklists and 2) compare the reporting quality of published vs. preprint studies.
Based on a search of the literature, 19 studies were selected for analysis (9 published studies and 10 preprint studies). The reporting quality of the studies was evaluated using the IOM, PRISMA, and MOOSE checklists.
The reporting quality of the published and preprint studies was similar, and was inferior in quality to that described in similar studies on reporting quality of meta-analyses published in the pathology and medicine literature.
Readers of COVID-19 laboratory hematology meta-analyses should be cognizant of their reporting quality problems, and critically evaluate them before using their information for patient care.
The issue of reporting quality is of critical importance, and the assessment of reporting quality has been underreported in the medical literature. Studies similar to this one will emphasize that the use of the IOM, PRISMA, and MOOSE checklists is a simple strategy to optimize the overall quality of meta-analyses.
Manuscript source: Invited manuscript
Specialty type: Hematology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Syngal S, Trivedi P, Wang DW S-Editor: Zhang H L-Editor: A P-Editor: Li JH
| 1. | Kinzler M, Zhang L. Underutilization of Meta-analysis in Diagnostic Pathology. Arch Pathol Lab Med. 2015;139:1302-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 2. | Eden J, Levit L, Berg L, Morton S. Finding what works in healthcare: standards for systematic reviews. Wsahington, DC.: The National Academies Press; 2011. |
| 3. | Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18665] [Cited by in RCA: 17454] [Article Influence: 1090.9] [Reference Citation Analysis (1)] |
| 4. | Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008-2012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14425] [Cited by in RCA: 16717] [Article Influence: 668.7] [Reference Citation Analysis (0)] |
| 5. | Liu X, Kinzler M, Yuan J, He G, Zhang L. Low Reporting Quality of the Meta-Analyses in Diagnostic Pathology. Arch Pathol Lab Med. 2017;141:423-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J Med Virol. 2020;92:401-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1670] [Cited by in RCA: 1766] [Article Influence: 353.2] [Reference Citation Analysis (0)] |
| 7. | Perlman S. Another Decade, Another Coronavirus. N Engl J Med. 2020;382:760-762. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 584] [Cited by in RCA: 517] [Article Influence: 103.4] [Reference Citation Analysis (0)] |
| 8. | Frater JL, Zini G, d'Onofrio G, Rogers HJ. COVID-19 and the clinical hematology laboratory. Int J Lab Hematol. 2020;42 Suppl 1:11-18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 154] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 9. | Lippi G, Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med. 2020;58:1131-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 502] [Cited by in RCA: 554] [Article Influence: 110.8] [Reference Citation Analysis (0)] |
| 10. | Lippi G, Plebani M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin Chem Lab Med. 2020;58:1063-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 198] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 11. | Borges do Nascimento IJ, Cacic N, Abdulazeem HM, von Groote TC, Jayarajah U, Weerasekara I, Esfahani MA, Civile VT, Marusic A, Jeroncic A, Carvas Junior N, Pericic TP, Zakarija-Grkovic I, Meirelles Guimarães SM, Luigi Bragazzi N, Bjorklund M, Sofi-Mahmudi A, Altujjar M, Tian M, Arcani DMC, O'Mathúna DP, Marcolino MS. Novel Coronavirus Infection (COVID-19) in Humans: A Scoping Review and Meta-Analysis. J Clin Med. 2020;9:941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 367] [Cited by in RCA: 326] [Article Influence: 65.2] [Reference Citation Analysis (2)] |
| 12. | Cao Y, Liu X, Xiong L, Cai K. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2: A systematic review and meta-analysis. J Med Virol. 2020; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 229] [Cited by in RCA: 208] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 13. | Fu L, Wang B, Yuan T, Chen X, Ao Y, Fitzpatrick T, Li P, Zhou Y, Lin YF, Duan Q, Luo G, Fan S, Lu Y, Feng A, Zhan Y, Liang B, Cai W, Zhang L, Du X, Li L, Shu Y, Zou H. Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: A systematic review and meta-analysis. J Infect. 2020;80:656-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 709] [Cited by in RCA: 640] [Article Influence: 128.0] [Reference Citation Analysis (0)] |
| 14. | Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020;58:1021-1028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 980] [Cited by in RCA: 1160] [Article Influence: 232.0] [Reference Citation Analysis (0)] |
| 15. | Lagunas-Rangel FA. Neutrophil-to-lymphocyte ratio and lymphocyte-to-C-reactive protein ratio in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. J Med Virol. 2020; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 375] [Cited by in RCA: 378] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 16. | Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clin Chim Acta. 2020;506:145-148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 946] [Cited by in RCA: 1129] [Article Influence: 225.8] [Reference Citation Analysis (0)] |
| 17. | Rodriguez-Morales AJ, Cardona-Ospina JA, Gutiérrez-Ocampo E, Villamizar-Peña R, Holguin-Rivera Y, Escalera-Antezana JP, Alvarado-Arnez LE, Bonilla-Aldana DK, Franco-Paredes C, Henao-Martinez AF, Paniz-Mondolfi A, Lagos-Grisales GJ, Ramírez-Vallejo E, Suárez JA, Zambrano LI, Villamil-Gómez WE, Balbin-Ramon GJ, Rabaan AA, Harapan H, Dhama K, Nishiura H, Kataoka H, Ahmad T, Sah R; Latin American Network of Coronavirus Disease 2019-COVID-19 Research (LANCOVID-19). Electronic address: https://www.lancovid.org. Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Med Infect Dis. 2020;34:101623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1647] [Cited by in RCA: 1431] [Article Influence: 286.2] [Reference Citation Analysis (0)] |
| 18. | Zhu J, Ji P, Pang J, Zhong Z, Li H, He C, Zhang J, Zhao C. Clinical characteristics of 3062 COVID-19 patients: A meta-analysis. J Med Virol. 2020; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 422] [Cited by in RCA: 375] [Article Influence: 75.0] [Reference Citation Analysis (0)] |
| 19. | Li LQ, Huang T, Wang YQ, Wang ZP, Liang Y, Huang TB, Zhang HY, Sun W, Wang Y. COVID-19 patients' clinical characteristics, discharge rate, and fatality rate of meta-analysis. J Med Virol. 2020;92:577-583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 886] [Cited by in RCA: 846] [Article Influence: 169.2] [Reference Citation Analysis (0)] |
| 20. | Arabi S, Vaseghi G, Heidari Z, Shariati L, Amin B, Rashid H, Javanmard SH. Clinical characteristics of COVID-19 infection in pregnant women: a systematic review and meta-analysis. MedRxiv. 2020;Preprint. [DOI] [Full Text] |
| 21. | Ebrahimi M, Malehi AS, Rahim F. Laboratory findings, signs and symptoms, clinical outcomes of Patients with COVID-19 Infection: an updated systematic review and meta-analysis. MedRxiv. 2020;Preprint. [DOI] [Full Text] |
| 22. | Han P, Han P, Diao K, Pang T, Huang S, Yang Z. Comparison of clinical features between critically and non-critically ill patients in SARS and COVID-19: a systematic review and meta-analysis. MedRxiv. 2020;Preprint. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 23. | Heydari K, Rismantab S, Shamshirian A, Lotfi P, Shadmehri N, Houshmand P, Zahedi M, Shamshirian D, Bathaeian S, Alizadeh-Navaei R. Clinical and Paraclinical Characteristics of COVID-19 patients: A systematic review and meta-analysis. MedRxiv. 2020;Preprint. [DOI] [Full Text] |
| 24. | Ma C, Gu J, Hou P, Zhang L, Bai Y, Guo Z, Wu H, Zhang B, Li P, Zhao X. Incidence, clinical characteristics and prognostic factor of patients with COVID-19: a systematic review and meta-analysis. MedRxiv. 2020;Preprint. [DOI] [Full Text] |
| 25. | Nasiri MJ, Haddadi S, Tahvildari A, Farsi Y, Arbabi M, Hasanzadeh S, Jamshidi P, Murthi M, Mirsaeidi MS. COVID-19 clinical characteristics, and sex-specific risk of mortality: Systematic Review and Meta-analysis. MedRxiv. 2020;Preprint. [DOI] [Full Text] |
| 26. | Pormohammad A, Ghorbani S, Khatami A, Farzi R, Baradaran B, Turner DL, Turner RJ, Bahr NC, Idrovo JP. Comparison of confirmed COVID-19 with SARS and MERS cases - Clinical characteristics, laboratory findings, radiographic signs and outcomes: A systematic review and meta-analysis. Rev Med Virol. 2020;30:e2112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 27. | Xu L, Mao Y, Chen G. Risk factors for severe corona virus disease 2019 (COVID-19) patients: a systematic review and meta analysis. MedRxiv. 2020;Preprint. [DOI] [Full Text] |
| 28. | Zhang HY, Jiao F, Wu X, Shang M, Luo Y, Gong Z. Clinical features, treatments and outcomes of severe and critical severe patients infected with COVID-19: A system review and meta-analysis. 2020; Preprint. [DOI] [Full Text] |
| 29. | Carlin JB. Tutorial in biostatistics. Meta-analysis: formulating, evaluating, combining, and reporting by S-L. T. Normand, Statistics in Medicine, 18, 321-359 (1999). Stat Med. 2000;19:753-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 30. | Andrews JC, Schünemann HJ, Oxman AD, Pottie K, Meerpohl JJ, Coello PA, Rind D, Montori VM, Brito JP, Norris S, Elbarbary M, Post P, Nasser M, Shukla V, Jaeschke R, Brozek J, Djulbegovic B, Guyatt G. GRADE guidelines: 15. Going from evidence to recommendation-determinants of a recommendation's direction and strength. J Clin Epidemiol. 2013;66:726-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 656] [Cited by in RCA: 962] [Article Influence: 80.2] [Reference Citation Analysis (0)] |
| 31. | Fleming PS, Koletsi D, Pandis N. Blinded by PRISMA: are systematic reviewers focusing on PRISMA and ignoring other guidelines? PLoS One. 2014;9:e96407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 32. | van Zuuren EJ, Fedorowicz Z. Moose on the loose: checklist for meta-analyses of observational studies. Br J Dermatol. 2016;175:853-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM; QUADAS-2 Group. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6953] [Cited by in RCA: 9480] [Article Influence: 677.1] [Reference Citation Analysis (0)] |
| 34. | Kelly J, Sadeghieh T, Adeli K. Peer Review in Scientific Publications: Benefits, Critiques, & A Survival Guide. EJIFCC. 2014;25:227-243. [PubMed] |
| 35. | Carneiroa CFD, Queiroza VGS, Moulina TC, Carvalhob C, Haase CB, Rayêef D, Henshallg DE, De-Souzaa EA, Amorima FE, Boosh FZ, Guercioi GD, Costaa IR, Hajduf KL, van Egmondj L, Modrákk M, Tanf PB, R.J. A, Burgessm SJ, Guerrad SFS, Bortoluzzie VT, Amarala OB. Comparing quality of reporting between preprints and peer-reviewed articles in the biomedical literature. 2020;Preprint. [DOI] [Full Text] |