Published online Aug 28, 2022. doi: 10.13105/wjma.v10.i4.195
Peer-review started: April 1, 2022
First decision: June 16, 2022
Revised: June 23, 2022
Accepted: July 27, 2022
Article in press: July 27, 2022
Published online: August 28, 2022
Processing time: 146 Days and 14.3 Hours
Due to the disease's broad clinical spectrum, it is currently unclear how to predict the future prognosis of patients at the time of diagnosis of coronavirus disease 2019 (COVID-19). Real-time reverse transcription-polymerase chain reaction (RT-PCR) is the gold standard molecular technique for diagnosing COVID-19. The number of amplification cycles necessary for the target genes to surpass a threshold level is represented by the RT-PCR cycle threshold (Ct) values. Ct values were thought to be an adequate proxy for severe acute respiratory syn
Core Tip: Real-time reverse transcription-polymerase chain reaction is regarded as the gold standard confirmatory test for coronavirus disease 2019 (COVID-19). Cycle threshold (Ct) values can be used to diagnose or forecast severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection since they are associated with virus load. Numerous differences exist in several clinical trials with small or large sample sizes, indicating a substantial positive correlation between the Ct value and disease severity in COVID-19. In this context, a literature review was conducted to address information gaps about the relationship between Ct levels and disease severity in COVID-19 patients globally. The majority of the data indicated a link between SARS-CoV-2 viral load and disease severity in these patients, and early anti-viral therapy will minimize the severity of the clinical outcomes.
- Citation: Leowattana W, Leowattana T, Leowattana P. SARS-CoV-2 viral load in the upper respiratory tract and disease severity in COVID-19 patients. World J Meta-Anal 2022; 10(4): 195-205
- URL: https://www.wjgnet.com/2308-3840/full/v10/i4/195.htm
- DOI: https://dx.doi.org/10.13105/wjma.v10.i4.195
Prior to November 2019, six coronaviruses (CoVs) were known to infect humans and cause respiratory disease: OC43, 229E, HKU1, and NL63, four community-acquired CoVs that are endemic in humans, as well as severe acute respiratory syndrome CoV (SARS-CoV) and Middle East respiratory syndrome CoV (MERS-CoV), two highly pathogenic CoVs that have zoonotic transmission followed by variable transmission between humans[1-5]. A new CoV discovered in late 2019 in Wuhan, Hubei Province, China has recently spread worldwide, causing a serious pandemic. SARS-CoV-2 was the name given to the new CoV, and the condition was dubbed coronavirus disease 2019 (COVID-19). SARS-CoV-2 spread rapidly from person to person, resulting in a pandemic that affected every province in China and eve
Viral load is used to diagnose severe viral infections of the respiratory system, as well as to track disease progression and treatment. By evaluating the value of the cycle threshold (Ct) of reverse transcription-polymerase chain reaction (RT-PCR), the SARS-CoV-2 viral load may be determined from the patient's viral RNA at a certain concentration. The lower the Ct values, the greater the viral load[9]. In contrast to other viral infections, no pathogen-specific prognostic indicators for SARS-CoV-2 are readily accessible. The first prognostic evaluation of individuals infected with SARS-CoV-2 may benefit from viral biomarkers capable of forecasting COVID-19 development in addition to the existing risk factors for severity. It is presently disputed whether the SARS-CoV-2 viral load affects the severity and course of the disease in this regard[10-13]. According to recent research, there may be a correlation between viral load and the severity of SARS-CoV-2 pneumonia, the degree of hypoxemia, the risk of mortality, as well as hematological, biochemical, and inflammatory alterations. However, diverse recruiting criteria have made it difficult to obtain a final, definite conclusion on the association between early nasopharyngeal viral load and individual outcomes[14-16]. The goal of this review is to ascertain if the SARS-CoV-2 Ct at diagnosis could anticipate the severity of COVID-19 and the outcomes of these patients.
Knudtzen et al[17] conducted a prospective cohort study of adult COVID-19 patients with PCR-positive SARS-CoV-2 airway samples to determine the association between cycle quantification (Cq)-values, hospitalization, and disease severity in 87 outpatients and 82 inpatients. The findings revealed that 31 of the 82 hospitalized patients (38.0%) had severe COVID-19 disease and had considerably lower baseline Cq-values than patients with moderate disease severity (median Cq-values = 24.8 vs 28.1, P = 0.01). They also discovered a statistically significant link between lower Cq-values and a higher risk of severe disease outcome (odds ratio [OR] = 0.89, 95% confidence interval (CI): 0.81-0.98, P = 0.018), which was independent of the timing of the test in relation to symptom onset and the presence of confounding factors such as airway sample type. When the date of the test and confounding variables were controlled for, they observed no relationship between lower baseline Cq-values in outpatients infected with SARS-CoV-2 and a greater likelihood of hospitalization. They concluded that SARS-CoV-2 PCR Cq-values were correlated with the time since symptoms began. Early in the clinical course, Cq-values were low as a sign of high viral loads, but Cq-values were not shown to be a predictor of hospitalization. On the other hand, lower Cq-values were found to be indicative of more disease severity in hospitalized patients.
Kawasuji et al[18] performed a retrospective cohort study to investigate the concentrations of SARS-CoV-2 RNA in the blood (RNAemia) and in the nasopharyngeal cavity, as well as their relationship with clinical severity in 56 COVID-19 patients. On admission, 19.6% (11/56) of patients had RNAemia, followed by 1.0% (1/25), 50.0% (6/12), and 100.0% (4/4) of intermediate, severe, and critically ill patients, respectively. Patients with RNAemia required more frequent oxygen supplementation (90.0% vs 13.3%), intensive care unit (ICU) admission (81.8% vs 6.7%), and invasive mechanical ventilation (27.3% vs 0.0%). The median viral load of nasopharyngeal swabs in patients with RNAemia was significantly higher in critically ill patients (5.4 Log10 copies/µL) than in moderate-severe cases (2.6 Log10 copies/µL), and significantly higher in non-survivors (6.2 Log10 copies/µL) than in survivors (3.9 Log10 copies/µL). They discovered a significant percentage of patients with SARS-CoV-2 RNAemia and a relationship between RNAemia and disease severity. Furthermore, among RNAemia patients, the viral loads of nasopharyngeal swabs were correlated with disease severity and death, suggesting the possibility of combining serum testing with nasopharyngeal tests as a prognostic indicator for COVID-19, with better quality than each test.
The connection between nasopharyngeal viral loads, host variables, and illness severity in 1122 SARS-CoV-2-infected patients was examined by Maltezou et al[19]. There were 309 (27.5%) patients with a high viral load, 316 (28.2%) with a moderate viral load, and 497 (44.3%) with a low viral load. In univariate analysis, individuals with high viral loads were older, had more comorbid diseases, required intubation for symptomatic disorders, and eventually passed away. Patients with a high viral load spent more time in the critical care unit and required more intubation than patients with a low viral load. Furthermore, individuals with chronic cardiovascular disease, hypertension, chronic lung disease, immunosuppression, obesity, and chronic neurological disease were more likely to have high viral loads. They concluded that viral load in the nasopharynx may be used to identify patients at high risk of morbidity or poor outcome.
Zheng et al[20] conducted a retrospective cohort study on 96 consecutively hospitalized COVID-19 patients, including 22 with moderate disease and 74 with severe disease, to assess viral loads at various phases of disease progression. After admission, 3497 respiratory, stool, serum, and urine samples were obtained from patients and tested for SARS-CoV-2 RNA viral load. RNA was also found in the feces of 55 (59%) of the patients and the serum of 39 (41%) of the patients. One patient's urine sample tested positive for SARS-CoV-2. The median duration of the virus in feces (22 d) was substantially longer than that in respiratory (18 d) and serum samples (16 d). Furthermore, the median duration of the virus in patients with severe disease (21 d) was substantially longer than that in patients with moderate disease (14 d). In the moderate group, viral loads peaked in respiratory samples in the second week after the illness started, but in the severe group, viral loads remained high throughout the third week. Virus duration was greater in individuals over the age of 60 and in men. They proposed that the duration of SARS-CoV-2 RNA in stool samples is significantly longer than that in respiratory and serum samples, emphasizing the importance of improving stool sample management in epidemic prevention and control and that the virus persists longer with higher load and peaks later in the respiratory tissue of patients with severe disease.
Aydin et al[21] investigated the predictive significance of viral load identified in the saliva of 300 COVID-19 patients in the early stages of illness. The results showed a mean Ct-value of 25.30 in the mild illness group, 19.85 in the intermediate disease group, 16.75 in the severe disease group, and 15.48 in the critical disease group. The pattern of the mean Ct-value of the oro-nasopharyngeal swab was similar to that of saliva. The authors concluded that the Ct-value of saliva and oro-nasopharyngeal swab might be used to predict disease severity.
de la Calle et al[14] performed a retrospective study of 455 hospitalized patients with a confirmed diagnosis of SARS-CoV-2 infection using prospective computerized medical data. The study population was separated into three groups based on the Ct value obtained upon admission: Patients with high viral load (Ct < 25), those with intermediate viral load (Ct 25-30), and those with low viral load (Ct > 30). The researchers discovered that 130 (28.6%) patients had a high viral load, 175 (38.5%) had an intermediate viral load, and 150 (33%) had a low viral load. They discovered that 120 (26.4%) patients died while they were in the hospital, and that 161 (35.4%) patients experienced respiratory failure after spending a median of 9 d there. High viral loads were associated with increased respiratory failure and a higher mortality rate at 30 d following admission in these patients. However, the risk of ICU admission was greater among patients with low and intermediate viral loads (12.3% vs 6.2%, P = 0.054). Septic shock, acute renal damage, venous thrombosis, hepatitis, or major adverse cardiovascular events were not different across groups. According to the authors, a useful prognostic indicator for the beginning of respiratory failure is the Ct value of RT-PCR in nasopharyngeal swabs at the time of admission.
Kwon et al[22] conducted a study on 31 hospitalized COVID-19 patients to investigate viral load, antibody responses to SARS-CoV-2, and cytokines/chemokines along the illness course, as well as to find parameters linked to disease severity. Asymptomatic and moderate patients had lower viral loads and longer viral shed than severe and critical cases. Unlike plasma IgG, which grew gradually and remained stable during hospitalization, plasma IgM peaked 3 wk after symptoms started and then declined. The antibody response was somewhat delayed but greater in severe and critical cases than in others. High levels of interferon (IFN)-α, IFN-γ induced protein-10, chemokine generated by IFN-γ, and interleukin-6 were linked with the severity of COVID-19 5-10 d after symptom onset. The authors hypothesized that a high viral load in the respiratory tract, as well as excessive cytokine and chemokine production between 1 and 2 wk after the onset of symptoms, was substantially linked with the severity of COVID-19.
Piubelli et al[23] conducted a retrospective analysis to assess the viral load of 373 confirmed COVID-19 patients seen in the emergency department between March 1, 2020 and May 31, 2020. According to the authors, 281 COVID-19 individuals were identified in March, 86 in April, and 6 in May. Along with a decline in the number of cases, they observed a considerable fall in the proportion of patients requiring critical care, which fell from 6.7% (19/281) in March to 1.1% (1/86) in April, and to none in May. In terms of viral load, they noticed a tendency for Ct to rise from a median of 24 to 34 between March and May, particularly in non-ICU patients. They concluded that throughout the pandemic, they saw a dramatic decline in severe COVID-19 patients that required critical care in addition to the declining viral load.
Shlomai et al[13] studied 170 hospitalized COVID-19 patients to see if there was a link between viral load at the time of admission, lung inflammation, and disease prognosis. The authors discovered that non-survivors and mechanically ventilated patients (n = 21) had a considerably greater virus load (8-fold, Ct = 23.43, P = 0.0001) than surviving non-intubated patients (n = 149, Ct = 29.55, P = 0.0001). Furthermore, a multivariate study adjusted for age, gender, and blood oxygen saturation (BOS)min found that low viral load was linked with a lower risk of mechanical ventilation and death (OR = 0.90, 95%CI: 0.81-0.99, P = 0.046). Furthermore, both BOS and patient age were independently related to mechanical ventilation and mortality (OR = 0.91, 95%CI: 0.84-0.98, P = 0.009 for BOS and OR = 1.05, 95%CI: 1.004-1.097 for patient age). They concluded that their data indicated a strong link between nasopharyngeal viral load and hypoxemia, as well as worse clinical outcomes in COVID-19 patients hospitalized.
In a study of 448 COVID-19 patients, Soria et al[24] looked at the relationship between viral load, as measured using nasopharyngeal swabs, and the severity of the illness. They clinically categorized individuals as having mild, moderate, or severe COVID-19 based on a variety of clinical characteristics such as the need for hospitalization, the necessity for oxygen treatment, admission to critical care units, and/or mortality. The authors discovered a statistically significant relationship between viral load and disease severity, with higher viral load associated with a worse clinical prognosis, independent of several previously identified risk factors such as age, gender, hypertension, cardiovascular diseases, diabetes, obesity, and pulmonary diseases.
Trunfio et al[25] conducted a study on 200 confirmed COVID-19 patients to see if the SARS-CoV-2 Ct value at diagnosis might predict COVID-19 disease severity, clinical symptoms, and 6-mo sequelae. Patients were divided into three groups based on diagnostic Ct values discovered from the initial swab: Ct 20, Group A; Ct = 20 - 28, Group B; and Ct > 28, Group C. The severity of the disease was graded on a six-point scale: Death, hospitalization with intubation, hospitalization needing continuous positive airway pressure support, hospitalization requiring low-flow wall oxygen to reservoir mask assistance, hospitalization without oxygen support, and no hospitalization. There were 168 survivors and 32 deaths among the 200 individuals. The range for the median age was 43-69. There were 116 (58.0%) men, and 188 of them were of European descent (94.0%). Patients with SARS-CoV-2 Ct were distributed as follows: 55 in Group A (27.5%), 55 in Group B (27.5%), and 90 in Group C (45.0%). Even after controlling for the time from COVID-19 onset to swab collection, the linear Ct values were negatively associated with the number of comorbidities per patient. Hospitalization-related patients were seen in Group A more frequently than in Group C (74.5% vs 56.7%). The severity of COVID-19 was substantially higher in Group A than in Groups B and C. With respect to Ct, there was an inverse distribution in the five categories of illness severity. Finally, 6-mo results for COVID-19 were worse in Group A than in the other groups; only 29.1% of patients in Group A had fully recovered at this point, compared to 70.9% and 80.0% in Groups B and C, respectively. Furthermore, Group A had a greater fatality rate (36.4%) than the other groups (Group B had a 12.7% lethality rate and Group C had a 5.6% lethality rate). After controlling for confounding variables, in multivariate analysis, lower SARS-CoV-2 Ct levels were independently associated with a greater risk of COVID-19-related death, along with older age and more comorbidities. The authors showed a correlation between COVID-19-related deaths, disease severity, the number of signs and symptoms at diagnosis, and the persistence of sequelae at 6 mo in symptomatic hospitalized and non-hospitalized patients, and the Ct value detected in nasopharyngeal swabs collected within the first week of COVID-19 onset.
Tsukagoshi et al[26] conducted a study on 286 confirmed COVID-19 patients to assess the links between epidemiological data, viral load, and disease severity (15 fatal cases, 133 symptomatic cases, and 138 asymptomatic cases). Compared to the number of viral copies at the time of sample collection, fatal cases had 3.57 ± 4.70 × 109 copies/mL, symptomatic cases had 3.92 ± 1.60 × 108 copies/mL, and asymptomatic cases had 4.92 ± 1.48 × 107 copies/mL. These findings imply that the viral loads of fatal and symptomatic patients were greater than those of asymptomatic cases. According to the authors, a high viral load of SARS-CoV-2 in elderly patients at an early stage of the disease, particularly those with pneumonia symptoms, results in a bad prognosis. Therefore, in such circumstances, we should intervene early to avoid the condition’s progressing to a severe degree.
Wang et al[27] conducted a study on 12 seriously ill and 11 slightly ill COVID-19 patients to explore the immune response and its link with clinical outcomes. The rates of viral replication, neutralizing antibody responses, and cross-reactivity with other human respiratory CoVs were also examined for use in the diagnosis, prognosis, and epidemiological studies. All 23 patients provided 461 clinical samples (84 nasal swabs, 59 throat swabs, 36 sputum samples, 90 fecal samples, 79 urine tests, and 113 plasma samples), including 1 stomach biopsy. They discovered that the majority of patients with severe illness shed viral loads for up to 30-40 d after beginning, but the majority of slightly unwell individuals had no detectable viral loads 15 d after onset. The peak viral load differed significantly between severe and moderate patients. The viral loads in the respiratory samples were larger in the severe group than in the mild group, and they gradually decreased with time. The SARS-CoV-2 was mostly found in respiratory samples. However, in the majority of critically ill patients, feces remained positive for viral RNA for an extended period of time. IgM responses in patients with severe disease increased within 1 to 2 wk after beginning and were progressively reduced after 4 wk, but IgM responses in patients with moderate disease were substantially lower. The majority of the mildly unwell patients (8/11) did not develop substantial IgM antibodies throughout the disease course, demonstrating that the IgM diagnosis was not sensitive for mildly ill individuals. IgG responses appeared 10-15 d after the initiation. The majority of patients had high levels of IgG antibodies that lasted at least 6 wk.
Faíco-Filho et al[28] conducted a retrospective cohort analysis on 875 confirmed COVID-19 patients to assess the relationship between SARS-CoV viral load and mortality. Fifty percent (439/875) of these patients had mild disease, 30.4% (266/875) had moderate disease, and 19.5% (170/875) had severe disease. In these COVID-19 individuals, a Ct value of 25 indicated a high viral load, which was independently related to death. They concluded that the SARS-CoV-2 virus load at admission was independently linked with death among hospitalized COVID-19 patients.
Pérez-García et al[29] conducted a retrospective study of 255 SARS-CoV-2–infected patients to determine the viral RNA content and expression of selected immune genes in the upper respiratory tract (nasopharynx), as well as their correlation with severe COVID-19. In the beginning, patients were split into three groups based on severity: 85 outpatients who underwent emergency room examinations and were discharged within the first 24 h (mild cases), 87 inpatients in medicine wards who did not require critical care (moderate cases), and 83 critical patients who were admitted to the ICU, or who passed away within 28 d of hospital admission (severe cases), and 30 healthy individuals were used as the control group. Interferon-stimulated gene 15 (ISG15), interferon-β (IFN-β), interferon-induced protein with tetratricopeptide repeats 1 (IFIT1), retinoic acid-inducible gene I (RIGI), tumor necrosis factor (TNF-β), interleukin 6 (IL-6), and chemokine (C-C motif) ligand 5 (CCL5) were all expressed at higher levels in COVID-19 patients. Individuals with severe COVID-19 had considerably greater SARS-CoV-2 viral load, IFN-β, IFIT1, IL-6, and IL-8 levels than patients with mild or moderate illness, although CCL5 values were significantly lower. They also found that ISG15, RIGI, TNF-β, IL-6, and CXCL10 strongly correlated with SARS-CoV-2 virus load. In adjusted regression models, SARS-CoV-2 viral load was a risk factor, but CCL5 was a protective factor for ICU admission or mortality during hospitalization. They also discovered significant relationships between the SARS-CoV-2 viral load and CCL5 in both cohorts when the entire cohort was divided in half, demonstrating a strong correlation between the severity of COVID-19 and both high levels of SARS-CoV-2 virus load and low levels of CCL5 exp
Guo et al[30] studied the relationship between SARS-CoV-2 viral load and disease severity in 195 hospitalized COVID-19 patients. The differences in clinical characteristics across four groups (mild, moderate, severe, and critical) and two groups (severe vs non-severe) were analyzed using one-way ANOVA and the student's t-test, respectively. More severe patients appear to have the following characteristics: Older age, underlying diseases, higher maximum body temperature within 24 h of hospitalization, longer time for virus clearance, longer duration of fever, higher levels of plasma C-reactive protein, D-dimer, procalcitonin, and aspartate aminotransferase, increased white blood cell count, particularly neutrophils, lower lymphocyte count, and higher initial viral load.
Tanner et al[31] performed a study on 185 hospitalized COVID-19 patients to assess the relationship between Ct value at admission and patient outcome while carefully controlling for confounders. On univariate analysis, the authors discovered that the Ct value at presentation was related to the likelihood of both ICU admission and mortality. Furthermore, Ct values changed considerably by age, length of illness at presentation, and antibody status. In a multivariate analysis, the Ct value was associated with the likelihood of death but not ICU admission. The presence of neutralizing antibodies at the time of presentation was not linked with death or ICU admission. They concluded that the SARS-CoV-2 Ct value at admission was independently related to mortality when other characteristics were controlled for and that it may be utilized for risk stratification.
Berastegui-Cabrera et al[32] conducted a prospective multicenter cohort study in 72 COVID-19 patients to assess the relationship between SARS-CoV-2 RNAemia, and viral load in the nasopharyngeal swab, and an unfavorable outcome, defined as ICU admission and/or death. Nine (12.5%) patients were treated as outpatients following an evaluation in the emergency room, whereas 63 (87.5%) patients were admitted to the hospital. Eleven (15.3%) of the patients were found to have SARS-CoV-2 RNAemia, with ten of them being hospitalized. The median viral load in plasma for the 11 SARS-CoV-2 RNAemia patients was 2.88 log10 copies/mL, while the median viral load in nasopharyngeal swabs for the 72 patients was 6.98 log10 copies/mL. Additionally, patients with SARS-CoV-2 RNAemia required more invasive mechanical ventilation (36.4% vs 6.6%) and had higher ICU admission rates (45.50% vs 8.2%) and ARDS (54.5% vs 9.8%). SARS-CoV-2 RNAemia patients exhibited a greater death rate (36.4% vs 4.9%) and a poorer prognosis (63.6% vs 13.1%). The authors concluded that patients with severe chronic liver disease and solid organ transplantation are more likely to have SARS-CoV-2 RNAemia at the time of the initial emergency room evaluation. They also noted that this condition is not predicted by a viral load in the upper respiratory airways and is linked to a poor prognosis.
Karahasan Yagci et al[33] conducted a study on 730 RT-PCR-positive patients to assess the severity of chest computed tomography (CT). Of the 284 patients admitted to the hospital, 27 (9.5%) died. There were no Ct results in 236 (32.3%) of the patients, and 216 (91.5%) of them were outpatients. In hospitalized patients, the total severity score (TSS) was much greater; 5.3% experienced severe alterations. Outpatients had lower Ct values, indicating a greater viral load. In both groups, an inverse relationship between viral load and TSS was seen. The severity of Ct was associated with age, with older individuals having a greater TSS. The authors concluded that viral load was not a significant risk factor for hospitalization or fatality. Outpatients exhibited high levels of viruses in their nasopharynx, making them infectious to their contacts. The viral load is critical in diagnosing the early stages of COVID-19 in order to limit potential transmission, whereas chest CT can assist in identifying patients that require significant medical treatment.
Le Borgne et al[34] conducted a retrospective study on 287 individuals with a confirmed diagnosis of COVID-19 to evaluate the association between SARS-CoV-2 viral load and disease severity. Nearly half of them (50.5%) had a moderate form, while the remaining half (49.5%) had a severe form that required mechanical ventilators. At admission, the median (interquartile range) viral load in the first upper respiratory swab was 4.76 (3.29-6.06) log10 copies/mL. This viral load measurement did not differ by subgroup when comparing survivors and non-survivors. Furthermore, the authors discovered that measuring respiratory viral load did not predict in-hospital mortality or disease severity. They claimed that the respiratory viral load in the first nasopharyngeal swab obtained during emergency department care is neither a predictor of the severity of the infection nor of death from SARS-CoV-2. The number of underlying comorbidities, as well as the host response to this viral infection, may be more predictive of disease severity than the virus itself.
Hasanoglu et al[35] studied the viral loads in six different sample types (nasopharyngeal, oral cavity, saliva, rectal, urine, and blood) from 60 patients to determine the relationship between disease severity and SARS-CoV-2 viral load, as well as differences in viral loads between asymptomatic and symptomatic patients. The authors discovered that 15 (25%) of the patients were asymptomatic, whereas 45 (75%) were symptomatic. There was a substantial difference in the mean ages of asymptomatic and symptomatic individuals (26.4 and 36.4, respectively). Asymptomatic individuals' viral loads were found to be substantially greater than symptomatic patients’. With increasing age, viral load has demonstrated a substantial negative tendency. With increasing disease severity, there was a considerable drop in viral load.
Bakir et al[36] conducted a study on 158 confirmed COVID-19 patients to evaluate the link between SARS-CoV-2 viral load Ct values and pneumonia. The authors discovered pneumonia in 40.5% of the individuals who underwent chest CT. SARS-CoV-2 Ct value and nasopharyngeal samples were shown to have a poor but significant connection with chest CT score. There was no link identified between viral load Ct value and age, gender, or death. There was no statistically significant relationship between chest CT score and death. The authors noted that the quantity of SARS-CoV-2 viral load did not correlate with the severity of the pulmonary lesions shown on chest CT.
Ng et al[37] studied 351 people (138 confirmed COVID-19 patients and 213 SARS-CoV-2-negative patients) to see if there is a link between SARS-CoV-2 viral load and disease severity. They discovered that viral loads in more seriously ill hospitalized patients, including those in the intensive care unit, were not significantly different from those in outpatient clinics. According to the authors, there is no clear association between viral load and disease severity, and a suitable biomarker for disease severity is currently unavailable in clinical settings.
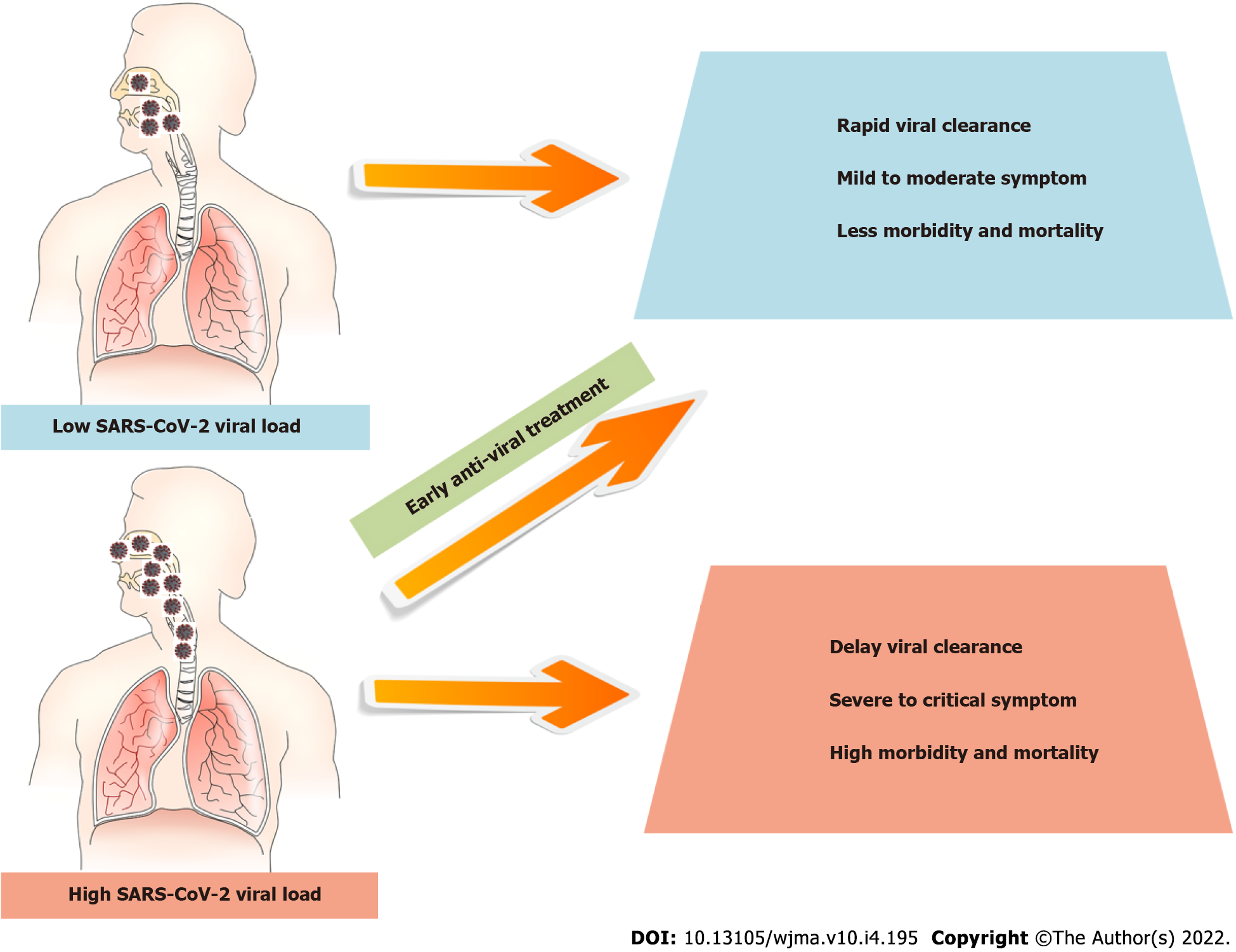
Although qualitative SARS-CoV-2 RT-PCR tests are routinely used to diagnose COVID-19, the therapeutic significance of quantitative information on Ct values being negatively associated with SARS-CoV-2 viral load for identifying viral copies must be understood. So far, several studies have shown inconsistent findings of the viral shedding kinetics in moderate and severe COVID-19 patients with an association or no association with disease severity. Table 1 summarizes the information regarding the countries of origin, study design, number of COVID-19 patients, mean or median Ct value, association of disease severity, and conclusions. The majority of the evidence suggests that a high SARS-CoV-2 viral load is associated with a severe clinical outcome. Along with this data, several studies found that patients admitted to the hospital with high SARS-CoV-2 virus loads, as determined by Ct values of nasopharyngeal swab samples, were more likely to be intubated or die during their hospitalization[11,16,38,39]. Furthermore, many researchers demonstrated that early antiviral treatment could effectively reduce virus load, shorten virus clearance time, and prevent COVID-19 from rapidly progressing to a severe disease outcome (Figure 1)[40-44].
| Ref. | Study design | No. of COVID-19 cases | Median/mean viral load (Ct or Cq) (log10 copies/mL) | Association with disease severity | Conclusion |
| Knudtzen et al[17], Denmark | Prospective cohort | 169 (87 OP/82 IP) | 24.8 vs 28.1 (Severe vs Moderate) | Yes | Lower Cq-values were found to be indicative of more disease severity in hospitalized patients |
| Kawasuji et al[18], Japan | Retrospective cohort | 56 (56 IP) | 5.4 vs 2.6 (Critical/Moderate-Severe) | Yes | The viral loads of NP swabs were correlated with disease severity and death |
| Maltezou et al[19], Greece | Prospective cohort | 1122 (274 OP/848 IP) | N/A | Yes | The viral load in the nasopharynx might be utilized to identify patients at increased risk for morbidity or poor outcome |
| Zheng et al[20], China | Retrospective cohort | 96 (96 IP) | N/A | Yes | The virus persists longer with higher load and peaks later in the respiratory tissue of patients with severe disease |
| Aydin et al[21], Turkey | Prospective cohort | 300 (168/79/29/24)(M/I/S/C) | 25.30/19.85/16.75/15.48 (M/I/S/C) | Yes | The Ct-values of saliva and oro-nasopharyngeal swab were useful in predicting disease severity |
| de la Calle et al[14], Spain | Retrospective cohort | 455 (455 IP) | N/A | Yes | The Ct value of RT-PCR in nasopharyngeal swabs on admission is a useful predictive marker for the development of respiratory failure |
| Kwon et al[22], Korea | Prospective cohort | 31 (31 IP) | 35.2/27.9/26.7 (M/I/S+C) | Yes | High viral load in the respiratory tract and excessive cytokines and chemokines were substantially linked with the severity of COVID-19 |
| Piubelli et al[23], Italy | Retrospective study | 373 (373 OP) | N/A | Yes | The decreasing viral load that they observed during March to May 2020 was associated with a significant reduction in severe COVID-19 cases that needed intensive care |
| Shlomai et al[13], Israel | Retrospective cohort | 170 (149 NS/21 SV) | 23.43 vs 29.55 (NS vs SV) | Yes | There was a clear relationship between nasopharyngeal viral load and hypoxemia, as well as worse clinical outcomes in hospitalized COVID-19 patients |
| Soria et al[24], Spain | Prospective cohort | 448 (110/236/102) (M/I/S) | 35.75/32.69/29.58 (M/I/S) | Yes | The link between viral load and disease severity was shown in COVID-19 patients |
| Trunfio et al[25], Italy | Retrospective cohort | 200 (32 NS/168 SV) | N/A | Yes | The Ct value detected within the first week of COVID-19 onset was associated with deaths and disease severity |
| Tsukagoshi et al[26], Japan | Retrospective study | 286 (138 AS/133 SM/15 FT) | N/A | Yes | The high viral load in elderly patients at an early stage of the disease results in a bad prognosis |
| Wang et al[27], China | Prospective cohort | 23 (11/12)(M/S) | N/A | Yes | The viral loads in the respiratory samples were larger in the severe group than in the mild group, and they gradually decreased with time |
| Faíco-Filho et al[28], Brazil | Retrospective cohort | 875 (439/266/170)(M/I/S) | 22/27/21.5 (M/I/S) | Yes | The SARS-CoV-2 virus load at admission was independently linked with death among hospitalized COVID-19 patients |
| Pérez-García et al[29], Spain | Retrospective study | 255 (85/87/83) (M/I/S) | N/A | Yes | The SARS-CoV-2 viral load was a risk factor, but CCL5 was a protective factor for ICU admission or mortality during hospitalization |
| Guo et al[30], China | Prospective cohort | 195 (16/132/41/6) (M/I/S/C) | 33.74/33.59/32.10/27.53 (M/I/S/C) | Yes | The higher initial viral load was associated with disease severity in COVID-19 patients |
| Tanner et al[31], United Kingdom | Prospective cohort | 185 (IP) | N/A | Yes | The SARS-CoV-2 Ct value at admission was independently related with mortality |
| Berastegui-Cabrera et al[32], Spain | Prospective cohort | 72 (9 OP/63 IP) | N/A | No | The viral load in the upper respiratory airways was associated with poor outcome |
| Karahasan Yagci et al[33], Turkey | Retrospective study | 730 (446 OP/284 IP) | (27.8/29.4/27.9) (M/I/S) | No | The viral load was not a significant risk factor for hospitalization or fatality |
| Le Borgne et al[34], France | Retrospective study | 287 (42 NS/245 SV) | 4.99 vs 4.76 (NS vs SV) | No | The viral load in the first nasopharyngeal swab was neither a predictor of severity nor of death in SARS-CoV-2 infection |
| Hasanoglu et al[35], Turkey | Prospective cohort | 60 (15 AS/45 SM) | N/A | No | Asymptomatic individuals' viral loads were found to be substantially greater than symptomatic patients' |
| Bakir et al[36], Turkey | Retrospective study | 158 (45 OP/113 IP) | 26.76 vs 27.53 (OP vs IP) | No | The quantity of SARS-CoV-2 viral load did not correlate with the severity of the pulmonary lesions shown on chest CT |
| Ng et al[37], USA | Retrospective study | 133 | N/A | No | The viral loads in more seriously ill hospitalized patients were not significantly different from those in outpatient clinics |
This review demonstrates an association between the Ct value discovered in nasopharyngeal swabs, which represented the quantitative SARS-CoV-2 viral load, and COVID-19-related fatalities and disease severity in both symptomatic hospitalized and non-hospitalized patients. These findings imply that the Ct value might be utilized as a tool to aid in the identification of individuals who are at a higher risk of having a catastrophic outcome. Early antiviral medication may successfully reduce viral load, decrease virus clearance time, and prevent the fast progression of COVID-19 to severe disease outcomes in this situation.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Infectious diseases
Country/Territory of origin: Thailand
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): E, E
P-Reviewer: Ata F, Qatar; Mohammadi M, Iran S-Editor: Liu JH L-Editor: Wang TQ P-Editor: Liu JH
| 1. | Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016;24:490-502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1725] [Cited by in RCA: 1876] [Article Influence: 208.4] [Reference Citation Analysis (0)] |
| 2. | Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181-192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3747] [Cited by in RCA: 3304] [Article Influence: 550.7] [Reference Citation Analysis (0)] |
| 3. | Gaunt ER, Hardie A, Claas EC, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48:2940-2947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 494] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 4. | Peiris JS, Yuen KY, Osterhaus AD, Stöhr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431-2441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 936] [Cited by in RCA: 900] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 5. | Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995-1007. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 781] [Cited by in RCA: 787] [Article Influence: 78.7] [Reference Citation Analysis (0)] |
| 6. | Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;55:105924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3736] [Cited by in RCA: 3197] [Article Influence: 639.4] [Reference Citation Analysis (0)] |
| 7. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14123] [Article Influence: 2824.6] [Reference Citation Analysis (1)] |
| 8. | European Centre for Disease Prevention and Control. COVID-19 situation update worldwide, as of 22 March 2022. Available from: https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases. |
| 9. | Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, Guo Q, Song T, He J, Yen HL, Peiris M, Wu J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382:1177-1179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3727] [Cited by in RCA: 3404] [Article Influence: 680.8] [Reference Citation Analysis (0)] |
| 10. | Rao SN, Manissero D, Steele VR, Pareja J. A Systematic Review of the Clinical Utility of Cycle Threshold Values in the Context of COVID-19. Infect Dis Ther. 2020;9:573-586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 282] [Cited by in RCA: 273] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 11. | Magleby R, Westblade LF, Trzebucki A, Simon MS, Rajan M, Park J, Goyal P, Safford MM, Satlin MJ. Impact of Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load on Risk of Intubation and Mortality Among Hospitalized Patients With Coronavirus Disease 2019. Clin Infect Dis. 2021;73:e4197-e4205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 256] [Cited by in RCA: 281] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 12. | Pujadas E, Chaudhry F, McBride R, Richter F, Zhao S, Wajnberg A, Nadkarni G, Glicksberg BS, Houldsworth J, Cordon-Cardo C. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8:e70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 317] [Cited by in RCA: 375] [Article Influence: 75.0] [Reference Citation Analysis (0)] |
| 13. | Shlomai A, Ben-Zvi H, Glusman Bendersky A, Shafran N, Goldberg E, Sklan EH. Nasopharyngeal viral load predicts hypoxemia and disease outcome in admitted COVID-19 patients. Crit Care. 2020;24:539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 14. | de la Calle C, Lalueza A, Mancheño-Losa M, Maestro-de la Calle G, Lora-Tamayo J, Arrieta E, García-Reyne A, Losada I, de Miguel B, Díaz-Simón R, López-Medrano F, Fernández-Ruiz M, Carretero O, San Juan R, Aguado JM, Lumbreras C. Impact of viral load at admission on the development of respiratory failure in hospitalized patients with SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis. 2021;40:1209-1216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 15. | Asai N, Sakanashi D, Ohashi W, Nakamura A, Yamada A, Kawamoto Y, Miyazaki N, Ohno T, Koita I, Suematsu H, Kishino T, Kato H, Hagihara M, Shiota A, Koizumi Y, Yamagishi Y, Mikamo H. Could threshold cycle value correctly reflect the severity of novel coronavirus disease 2019 (COVID-19)? J Infect Chemother. 2021;27:117-119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 16. | Argyropoulos KV, Serrano A, Hu J, Black M, Feng X, Shen G, Call M, Kim MJ, Lytle A, Belovarac B, Vougiouklakis T, Lin LH, Moran U, Heguy A, Troxel A, Snuderl M, Osman I, Cotzia P, Jour G. Association of Initial Viral Load in Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Patients with Outcome and Symptoms. Am J Pathol. 2020;190:1881-1887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 132] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 17. | Knudtzen FC, Jensen TG, Lindvig SO, Rasmussen LD, Madsen LW, Hoegh SV, Bek-Thomsen M, Laursen CB, Nielsen SL, Johansen IS. SARS-CoV-2 viral load as a predictor for disease severity in outpatients and hospitalised patients with COVID-19: A prospective cohort study. PLoS One. 2021;16:e0258421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 18. | Kawasuji H, Morinaga Y, Tani H, Yoshida Y, Takegoshi Y, Kaneda M, Murai Y, Kimoto K, Ueno A, Miyajima Y, Fukui Y, Kimura M, Yamada H, Sakamaki I, Yamamoto Y. SARS-CoV-2 RNAemia with a higher nasopharyngeal viral load is strongly associated with disease severity and mortality in patients with COVID-19. J Med Virol. 2022;94:147-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 19. | Maltezou HC, Raftopoulos V, Vorou R, Papadima K, Mellou K, Spanakis N, Kossyvakis A, Gioula G, Exindari M, Froukala E, Martinez-Gonzalez B, Panayiotakopoulos G, Papa A, Mentis A, Tsakris A. Association Between Upper Respiratory Tract Viral Load, Comorbidities, Disease Severity, and Outcome of Patients With SARS-CoV-2 Infection. J Infect Dis. 2021;223:1132-1138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 20. | Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, Xie G, Lin S, Wang R, Yang X, Chen W, Wang Q, Zhang D, Liu Y, Gong R, Ma Z, Lu S, Xiao Y, Gu Y, Zhang J, Yao H, Xu K, Lu X, Wei G, Zhou J, Fang Q, Cai H, Qiu Y, Sheng J, Chen Y, Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1052] [Cited by in RCA: 1022] [Article Influence: 204.4] [Reference Citation Analysis (0)] |
| 21. | Aydin S, Benk IG, Geckil AA. May viral load detected in saliva in the early stages of infection be a prognostic indicator in COVID-19 patients? J Virol Methods. 2021;294:114198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Kwon JS, Kim JY, Kim MC, Park SY, Kim BN, Bae S, Cha HH, Jung J, Kim MJ, Lee MJ, Choi SH, Chung JW, Shin EC, Kim SH. Factors of Severity in Patients with COVID-19: Cytokine/Chemokine Concentrations, Viral Load, and Antibody Responses. Am J Trop Med Hyg. 2020;103:2412-2418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 23. | Piubelli C, Deiana M, Pomari E, Silva R, Bisoffi Z, Formenti F, Perandin F, Gobbi F, Buonfrate D. Overall decrease in SARS-CoV-2 viral load and reduction in clinical burden: the experience of a hospital in northern Italy. Clin Microbiol Infect. 2021;27:131.e1-131.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Soria ME, Cortón M, Martínez-González B, Lobo-Vega R, Vázquez-Sirvent L, López-Rodríguez R, Almoguera B, Mahillo I, Mínguez P, Herrero A, Taracido JC, Macías-Valcayo A, Esteban J, Fernandez-Roblas R, Gadea I, Ruíz-Hornillos J, Ayuso C, Perales C. High SARS-CoV-2 viral load is associated with a worse clinical outcome of COVID-19 disease. Access Microbiol. 2021;3:000259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 25. | Trunfio M, Venuti F, Alladio F, Longo BM, Burdino E, Cerutti F, Ghisetti V, Bertucci R, Picco C, Bonora S, Di Perri G, Calcagno A. Diagnostic SARS-CoV-2 Cycle Threshold Value Predicts Disease Severity, Survival, and Six-Month Sequelae in COVID-19 Symptomatic Patients. Viruses. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 26. | Tsukagoshi H, Shinoda D, Saito M, Okayama K, Sada M, Kimura H, Saruki N. Relationships between Viral Load and the Clinical Course of COVID-19. Viruses. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 27. | Wang Y, Zhang L, Sang L, Ye F, Ruan S, Zhong B, Song T, Alshukairi AN, Chen R, Zhang Z, Gan M, Zhu A, Huang Y, Luo L, Mok CKP, Al Gethamy MM, Tan H, Li Z, Huang X, Li F, Sun J, Zhang Y, Wen L, Li Y, Chen Z, Zhuang Z, Zhuo J, Chen C, Kuang L, Wang J, Lv H, Jiang Y, Li M, Lin Y, Deng Y, Tang L, Liang J, Huang J, Perlman S, Zhong N, Zhao J, Malik Peiris JS. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest. 2020;130:5235-5244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 419] [Article Influence: 83.8] [Reference Citation Analysis (0)] |
| 28. | Faíco-Filho KS, Passarelli VC, Bellei N. Is Higher Viral Load in SARS-CoV-2 Associated with Death? Am J Trop Med Hyg. 2020;103:2019-2021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 29. | Pérez-García F, Martin-Vicente M, Rojas-García RL, Castilla-García L, Muñoz-Gomez MJ, Hervás Fernández I, González Ventosa V, Vidal-Alcántara EJ, Cuadros-González J, Bermejo-Martin JF, Resino S, Martínez I. High SARS-CoV-2 Viral Load and Low CCL5 Expression Levels in the Upper Respiratory Tract Are Associated With COVID-19 Severity. J Infect Dis. 2022;225:977-982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 30. | Guo X, Jie Y, Ye Y, Chen P, Li X, Gao Z, Li G, Deng H, Zheng Y, Lin B, Chong Y, Chen F. Upper Respiratory Tract Viral Ribonucleic Acid Load at Hospital Admission Is Associated With Coronavirus Disease 2019 Disease Severity. Open Forum Infect Dis. 2020;7:ofaa282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 31. | Tanner AR, Phan H, Brendish NJ, Borca F, Beard KR, Poole S, W Clark T. SARS-CoV-2 viral load at presentation to hospital is independently associated with the risk of death. J Infect. 2021;83:458-466. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Berastegui-Cabrera J, Salto-Alejandre S, Valerio M, Pérez-Palacios P, Revillas FAL, Abelenda-Alonso G, Oteo-Revuelta JA, Carretero-Ledesma M, Muñoz P, Pascual Á, Gozalo M, Rombauts A, Alba J, García-Díaz E, Rodríguez-Ferrero ML, Valiente A, Fariñas MC, Carratalà J, Santibáñez S, Camacho-Martínez P, Pachón J, Cisneros JM, Cordero E, Sánchez-Céspedes J. SARS-CoV-2 RNAemia is associated with severe chronic underlying diseases but not with nasopharyngeal viral load. J Infect. 2021;82:e38-e41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Karahasan Yagci A, Sarinoglu RC, Bilgin H, Yanılmaz Ö, Sayın E, Deniz G, Guncu MM, Doyuk Z, Barıs C, Kuzan BN, Aslan B, Korten V, Cimsit C. Relationship of the cycle threshold values of SARS-CoV-2 polymerase chain reaction and total severity score of computerized tomography in patients with COVID 19. Int J Infect Dis. 2020;101:160-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 34. | Le Borgne P, Solis M, Severac F, Merdji H, Ruch Y, Alamé Intern K, Bayle E, Hansmann Y, Bilbault P, Fafi-Kremer S, Meziani F; CRICS TRIGGERSEP Group (Clinical Research in Intensive Care and Sepsis Trial Group for Global Evaluation and Research in Sepsis). SARS-CoV-2 viral load in nasopharyngeal swabs in the emergency department does not predict COVID-19 severity and mortality. Acad Emerg Med. 2021;28:306-313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 35. | Hasanoglu I, Korukluoglu G, Asilturk D, Cosgun Y, Kalem AK, Altas AB, Kayaaslan B, Eser F, Kuzucu EA, Guner R. Higher viral loads in asymptomatic COVID-19 patients might be the invisible part of the iceberg. Infection. 2021;49:117-126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 73] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 36. | Bakir A, Hosbul T, Cuce F, Artuk C, Taskin G, Caglayan M, Guney M, Kurkcu MF, Yildiz F, Erdal H, Erdem G. Investigation of Viral Load Cycle Threshold Values in Patients with SARS-CoV-2 Associated Pneumonia with Real-Time PCR Method. J Infect Dev Ctries. 2021;15:1408-1414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 37. | Ng DL, Granados AC, Santos YA, Servellita V, Goldgof GM, Meydan C, Sotomayor-Gonzalez A, Levine AG, Balcerek J, Han LM, Akagi N, Truong K, Neumann NM, Nguyen DN, Bapat SP, Cheng J, Martin CS, Federman S, Foox J, Gopez A, Li T, Chan R, Chu CS, Wabl CA, Gliwa AS, Reyes K, Pan CY, Guevara H, Wadford D, Miller S, Mason CE, Chiu CY. A diagnostic host response biosignature for COVID-19 from RNA profiling of nasal swabs and blood. Sci Adv. 2021;7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 71] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 38. | La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, Gautret P, Raoult D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059-1061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 682] [Cited by in RCA: 690] [Article Influence: 138.0] [Reference Citation Analysis (0)] |
| 39. | Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4682] [Cited by in RCA: 4809] [Article Influence: 961.8] [Reference Citation Analysis (0)] |
| 40. | Yu T, Tian C, Chu S, Zhou H, Zhang Z, Luo S, Hu D, Fan H. COVID-19 patients benefit from early antiviral treatment: A comparative, retrospective study. J Med Virol. 2020;92:2675-2683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 41. | Çalık Başaran N, Uyaroğlu OA, Telli Dizman G, Özışık L, Şahin TK, Taş Z, İnkaya AÇ, Karahan S, Alp Ş, Alp A, Metan G, Zarakol P, Sain Güven G, Öz ŞG, Topeli A, Uzun Ö, Akova M, Ünal S. Outcome of noncritical COVID-19 patients with early hospitalization and early antiviral treatment outside the ICU. Turk J Med Sci. 2021;51:411-420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 42. | Wu J, Li W, Shi X, Chen Z, Jiang B, Liu J, Wang D, Liu C, Meng Y, Cui L, Yu J, Cao H, Li L. Early antiviral treatment contributes to alleviate the severity and improve the prognosis of patients with novel coronavirus disease (COVID-19). J Intern Med. 2020;288:128-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 218] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 43. | Karatas E, Aksoy L, Ozaslan E. Association of Early Favipiravir Use with Reduced COVID-19 Fatality among Hospitalized Patients. Infect Chemother. 2021;53:300-307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 44. | Vena A, Cenderello G, Balletto E, Mezzogori L, Santagostino Barbone A, Berruti M, Ball L, Battaglini D, Bonsignore A, Dentone C, Giacobbe DR, Eldin TK, Mikulska M, Rebesco B, Robba C, Scintu A, Stimamiglio A, Taramasso L, Pelosi P, Artioli S, Bassetti M. Early Administration of Bamlanivimab in Combination with Etesevimab Increases the Benefits of COVID-19 Treatment: Real-World Experience from the Liguria Region. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |