Published online Aug 16, 2023. doi: 10.12998/wjcc.v11.i23.5504
Peer-review started: May 19, 2023
First decision: June 21, 2023
Revised: June 29, 2023
Accepted: July 25, 2023
Article in press: July 25, 2023
Published online: August 16, 2023
Processing time: 88 Days and 23.1 Hours
The objectives of this study were to identify hub genes and biological pathways involved in lung adenocarcinoma (LUAD) via bioinformatics analysis, and investigate potential therapeutic targets.
To determine reliable prognostic biomarkers for early diagnosis and treatment of LUAD.
To identify potential therapeutic targets for LUAD, two microarray datasets derived from the Gene Expression Omnibus (GEO) database were analyzed, GSE3116959 and GSE118370. Differentially expressed genes (DEGs) in LUAD and normal tissues were identified using the GEO2R tool. The Hiplot database was then used to generate a volcanic map of the DEGs. Weighted gene co-expression network analysis was conducted to cluster the genes in GSE116959 and GSE
Three hub genes with high connectivity were identified; cellular retinoic acid binding protein 2 (CRABP2), matrix metallopeptidase 12 (MMP12), and DNA topoisomerase II alpha (TOP2A). High expression of these genes was associated with a poor LUAD prognosis, and the genes exhibited high diagnostic value.
Expression levels of CRABP2, MMP12, and TOP2A in LUAD were higher than those in normal lung tissue. This observation has diagnostic value, and is linked to poor LUAD prognosis. These genes may be biomarkers and therapeutic targets in LUAD, but further research is warranted to investigate their usefulness in these respects.
Core Tip: Lung cancer is an important cause of cancer-related death worldwide. This study conducted multiple bioinformatics analysis methods to explore potential therapeutic targets for lung adenocarcinoma (LUAD). Finally, three hub genes with high connectivity were identified, namely cellular retinoic acid binding protein 2, matrix metallopeptidase 12, and DNA topoisomerase II alpha. High expression of these genes was associated with a poor LUAD prognosis, and the genes exhibited high diagnostic value. Therefore, these genes may be biomarkers and therapeutic targets for LUAD.
- Citation: Zhang L, Liu Y, Zhuang JG, Guo J, Li YT, Dong Y, Song G. Identification of key genes and biological pathways in lung adenocarcinoma by integrated bioinformatics analysis. World J Clin Cases 2023; 11(23): 5504-5518
- URL: https://www.wjgnet.com/2307-8960/full/v11/i23/5504.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i23.5504
Lung cancer is an important cause of cancer-related death worldwide, with incidence and mortality rates of 11.6% and 18.4%, respectively[1,2]. Non-small cell lung cancer is the most common type of lung cancer, and lung adenocarcinoma (LUAD) is the main histological subtype of non-small cell lung cancer[3]. Most cases of LUAD are diagnosed at an advanced or metastatic stage; in addition, the 5-year survival rate is extremely low[4-6]. Currently, treatment effectiveness for advanced LUAD is limited. Moreover, LUAD is associated with a high risk of recurrence after treatment due to the heterogeneity of tumors[7]. Therefore, it is imperative to identify a reliable prognostic biomarker for early diagnosis and treatment. The use of such biomarkers could potentially improve the prognosis of patients with lung cancer.
The gene chip is a systematic high-throughput method for detecting and analyzing differentially expressed genes (DEGs) in various tissues. This technique can assist in the identification of prognostic genes and biomarkers of cancer[8-10]. Thus, it has become a powerful tool for studying the differential expression of genes related to the carcinogenesis and progression of LUAD. In addition, it provides the possibility to analyze the tumor microenvironment and the functional diversity of tumor-infiltrating immune cells[11-13]. With the rapid development and application of gene chip technology, researchers have accumulated a large amount of gene data. Hence, gene mining based on these data has become a research hotspot[14]. In this study, various bioinformatics methods were utilized to identify the core genes associated with the prognosis of LUAD. These core genes can be used for disease diagnosis and prognosis, and may help elucidate the mechanisms underlying LUAD.
The National Center for Biotechnology Information-Gene Expression Omnibus (NCBI-GEO) is a public database (https://www.ncbi.nlm.nih.gov/geo/). Using the keywords “Lung Adenocarcinoma” and “Homo Sapiens” in our search, we obtained two datasets, GSE116959 (including 57 LUAD tissues and 11 normal tissues) and GSE118370 (including 6 LUAD and 6 normal tissues). These datasets were downloaded and analyzed using GEO2R.
We analyzed DEGs between LUAD and normal tissues in the GSE116959 and GSE118370 datasets using the GEO2R tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/). Volcano maps and Wayne diagrams drawn for each dataset were obtained from the Hiplot database[15] (https://hiplot-academic.com/).
Weighted gene co-expression network analysis (WGCNA) is a systems biology method that can interconnect and cluster different types of genes into various modules to calculate the correlation between genes and clinical practice[16]. The online tool EHBIO platform (http://www.ehbio.com/Cloud_Platform/front/#/) was used to perform WGCNA on the GSE116959 and GSE118370 datasets. After screening, modules that were more relevant to the sample characteristics were included in subsequent analyses.
Database for Annotation, Visualization and Integrated Discovery (DAVID)[17] is a web server for gene lists, functional enrichment analysis, and functional annotation (https://david.ncifcrf.gov/). The latest version of the DAVID database (version 7.0) was used to conduct Gene Ontology (GO) enrichment analyses of the DEGs.
The Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string-db.org/) is designed to analyze protein-protein interaction (PPI) information. To evaluate potential PPI relationships, the previously identified DEGs were mapped to the STRING database. The PPI pairs with a combined score of 0.4 were extracted. Subsequently, the PPI network was visualized using the Cytoscape software (www.cytoscape.org/). Nodes with higher degree and higher betweenness centrality of connectivity tend to be more essential for maintaining the stability of the entire network. CytoHubba and CytoNCA are two plugins in Cytoscape that are used to calculate the degree and betweenness centrality of each protein node. In this study, genes with connectivity ≥ 10 and betweenness centrality ≥ 50 were identified as hub genes.
GraphPad Prism 8.0 software (GraphPad Software Inc., San Diego, CA, United States) was used to perform receiver operating characteristic (ROC) statistical analysis of the hub genes to evaluate their diagnostic value. The Kaplan-Meier plotter (http://kmplot.com/analysis/) mRNA lung cancer database was then used to evaluate the prognostic value of hub genes in patients with lung cancer. Patients with cancer were classified into two groups according to the median mRNA expression of each gene. P < 0.05 was deemed to indicate statistically significance.
The single-sample gene set enrichment analysis (ssGSEA) is an extension of the GSEA method used to calculate the enrichment score of each sample and gene set pairing. We conducted ssGSEA on the identified hub genes using the gene set variation analysis R package.
The University of Alabama at Birmingham Cancer data analysis portal (UALCAN)[18] is an interactive network resource (http://ualcan.path.uab.edu/). We analyzed the differential expression of hub genes in normal and tumor tissues, as well as their expression during tumor progression using relevant data from the UALCAN.
The Human Protein Atlas (HPA) database (https://www. Proteinatlas.org/) provides detailed information on the distribution of proteins in human tissues and cells. We selected immunohistochemical images of the hub genes in lung cancer and normal tissues from the HPA database. These images were used to detect the differential expression of hub genes at the protein level.
Table 1 shows the two datasets that were selected in this study. Based on the criteria (P < 0.05 and log fold change > 2.00), a total of 424 and 409 DEGs were identified from GSE116959 and GSE118370, respectively. The corresponding volcano maps for GSE116959 (Figure 1A) and GSE118370 (Figure 1B) are shown.
| Dataset ID | Tumor | Normal | Total number |
| GSE116959 | 57 | 11 | 68 |
| GSE118370 | 6 | 6 | 12 |
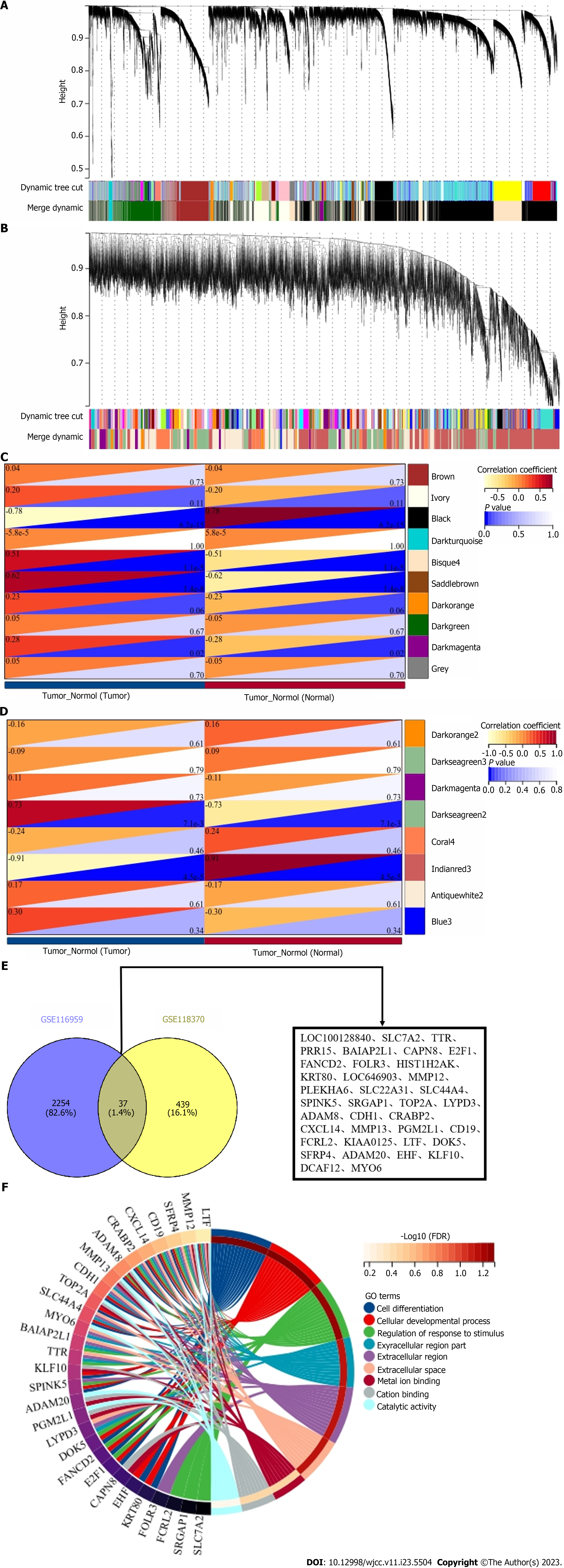
Using WGCNA, 10 modules were identified in GSE116959 (each color represents a module). Based on the Spearman correlation coefficient, a heat map of the module-trait relationship was generated to evaluate correlations between each module and the disease (Figure 2A and C). Four modules exhibited high correlations with LUAD; black (r = -0.78, P = 6.2e-12), disque4 (r = 0.51, P = 1.1e-8), saddle brown (r = 0.62, P = 1.4e-8), and dark magenta (r = 0.28, P = 0.02). Among them, disque4, saddle brown, and dark magenta were positively correlated with LUAD, including a total of 2291 DEGs.
Eight modules were identified in GSE118370 (Figure 2B and D). Two modules were highly correlated with LUAD, Indian red 3 (r = -0.91, P = 4.5e-5) and dark sea green 2 (r = 0.73, P = 7.1e-3). Among them, dark sea green 2 was positively correlated with LUAD, containing 476 DEGs in total. A cross analysis was conducted on two datasets, resulting in identification of a total of 37 shared DEGs (Figure 2E).
GO function enrichment analyses for DEGs were performed using the DAVID database. The enriched GO terms were divided into cellular component, molecular function, and biological process ontologies. The results indicated that, in the cellular component ontology, the DEGs were enriched in the extracellular region, extracellular region part, and extracellular space. In the molecular function ontology, the analysis showed that the DEGs were significantly enriched in metal ion binding, cation binding, and catalytic activity. In the biological process ontology, the DEGs were enriched in cell differentiation, cellular developmental process, and regulation of response to stimulus (Figure 2F).
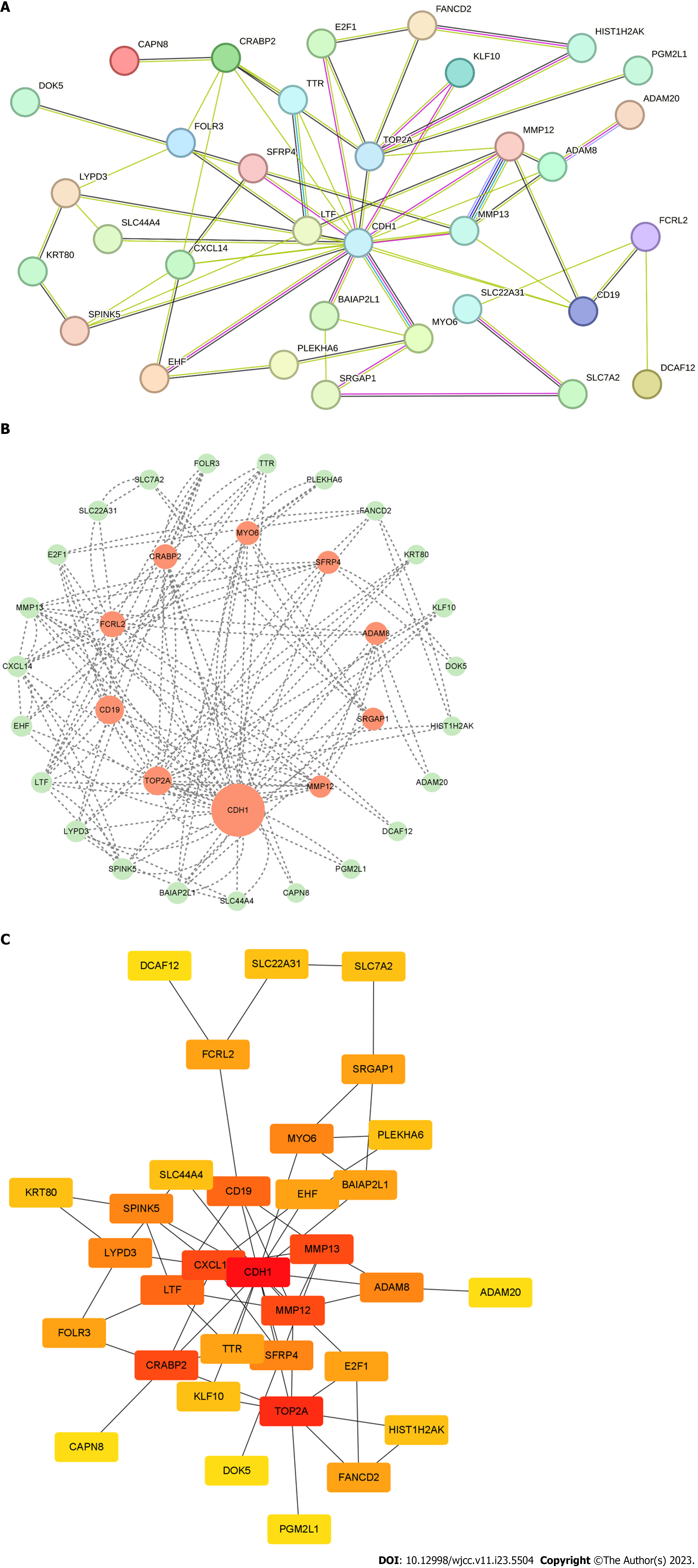
We constructed a PPI network which involved a total of 35 nodes and 61 edges (Figure 3A). Afterwards, the degree and betweenness centrality of each protein node were calculated (Figure 3B and C). The analysis identified a total of five genes; cadherin 1 (CDH1), CD19, CRABP2, matrix metallopeptidase 12 (MMP12), and DNA topoisomerase II alpha (TOP2A) (Table 2; Figure 4).
| Gene symbol | Betweenness centrality (≥ 50) | Gene symbol | Degree (≥ 10) | |
| 1 | CDH1 | 611.39685 | CDH1 | 34 |
| 2 | TOP2A | 167.13333 | TOP2A | 16 |
| 3 | CD19 | 166.32063 | CRABP2 | 12 |
| 4 | FCRL2 | 117.73333 | MMP13 | 12 |
| 5 | CRABP2 | 96.13333 | MMP12 | 12 |
| 6 | MYO6 | 78.28889 | CXCL14 | 12 |
| 7 | SFRP4 | 62.00000 | CD19 | 10 |
| 8 | ADAM8 | 62.00000 | LTF | 10 |
| 9 | SRGAP1 | 56.266666 | - | - |
| 10 | MMP12 | 51.720634 | - | - |
The diagnostic value of the hub genes was evaluated through ROC analysis of GSE116959 and GSE118370 (Figure 5A and B). The area under the ROC curve was > 75% for all five hub genes, indicating that these genes have strong diagnostic value. Subsequently, the prognostic value of the hub genes was evaluated using the Kaplan-Meier plotter bioinformatics analysis platform (Figure 5C-G). The results showed that an increase in the expression levels of CDH1 was significantly negatively correlated with the prognosis of lung cancer. In contrast, an increase in the expression levels of CRABP2, MMP12, and TOP2A was significantly positively correlated with the prognosis of lung cancer. The results indicate that CRABP2, MMP12, and TOP2A are associated with a poor LUAD prognosis.
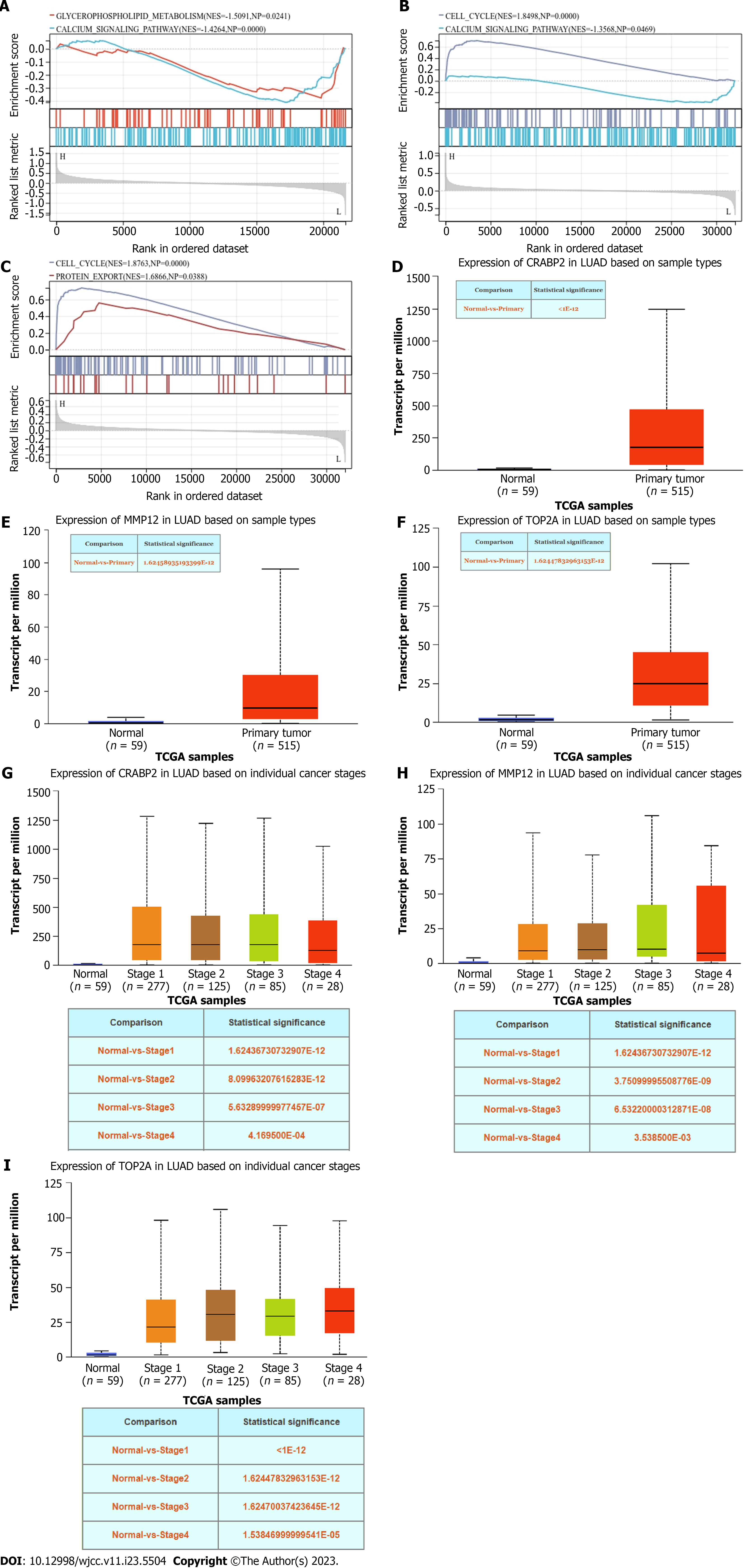
By performing ssGSEA on the hub genes, we found that CRABP2 mainly affected glycerophospholipid metabolism and the calcium signaling pathway. MMP12 mainly affected the cell cycle and calcium signaling pathway. TOP2A mainly affected the cell cycle and protein export (Figure 6A-C). Thereafter, we analyzed the hub genes using the UALCAN. The results showed that the expression of CRABP2, MMP12, and TOP2A in LUAD tissue was significantly increased in tumor tissues compared with normal samples (Figure 6D-F). We also found that the expression levels of CRABP2, MMP12, and TOP2A were significantly increased during the progression of tumors (Figure 6G-I).
The analysis of immunohistochemical images showed that the protein expression of CRABP2, MMP12, and TOP2A was significantly increased in LUAD compared with normal tissues (Figure 7).
In recent decades the incidence rate of LUAD has gradually increased, and it has become one of the most common types of lung cancer tumors. The prognosis of LUAD remains poor, and the 5-year relative survival rate is < 21%[19,20]. The treatment options for LUAD are currently limited to surgical resection and chemotherapy, both of which exhibit poor effectiveness[21,22]. The poor prognosis of LUAD is mainly attributed to the lack of specific biomarkers, which could permit early diagnosis and targeted treatment[23]. Therefore, the discovery of effective biomarkers has become a key factor in the treatment of LUAD. Bioinformatics is a new analytical method that differs from traditional experimental approaches. It is mainly used to process and analyze biological data via mathematical formulas and statistical methods[24]. Numerous studies have investigated biomarkers related to LUAD[25,26], but research conducted thus far has not identified effective biomarkers. It is therefore crucial to analyze LUAD from various aspects, identify the best target molecules for its treatment, and elucidate the biological pathways underlying its development and progression.
In the current study WGCNA revealed 37 shared DEGs in the two selected datasets. GO enrichment analysis indicated that the DEGs mainly pertained to immune-related mechanisms, including extracellular regions, extracellular regions, extracellular space, metal ion binding, cation binding, catalytic activity, cell differentiation, cellular developmental processes, and regulation of responses to stimuli. These data further indicated the molecular mechanisms associated with LUAD, and suggested that tumor pathogenesis is a complex biological process caused by changes in the expression of specific genes and epigenetic alterations. The abnormal regulation of multiple genes can promote the occurrence and development of LUAD via different pathways.
To further search for hub genes, a PPI network was constructed and analysis of each protein node was performed. Five genes satisfied the criteria; CDH 1, CD19, CRABP2, MMP12, and TOP2A. ROC analysis demonstrated that these five hub genes had high diagnostic value. The prognostic value of these five genes was then evaluated. Kaplan-Meier analysis indicated that increased expression of CRABP2, MMP12, and TOP2A can worsen LUAD, resulting in a poorer prognosis.
ssGSEA was conducted on the hub genes to investigate whether CRABP2, MMP12, and TOP2A affect LUAD. The results suggest that CRABP2 mainly affects glycerophospholipid metabolism and the calcium signaling pathway. MMP12 mainly affects the cell cycle and calcium signaling pathway. TOP2A mainly affects the cell cycle and protein export. Expression levels of CRABP2, MMP12, and TOP2A in LUAD were then investigated using the UALCAN. Compared with normal samples, CRABP2, MMP12, and TOP2A expression levels were significantly increased in LUAD tissue. Their expression was also significantly increased during tumor progression. These data indicate that CRABP2, MMP12, and TOP2A may lead to the occurrence and progression of LUAD, and have a sustained impact on LUAD. Lastly, immunohistochemical staining analysis revealed that protein expression of CRABP2, MMP12, and TOP2A in LUAD tissue was markedly higher than that in normal tissue. These findings further highlighted the importance of CRABP2, MMP12, and TOP2A in LUAD.
In summary, CRABP2, MMP12, and TOP2A play a major role in the initiation and progression of LUAD. The present study indicates that increased CRABP2, MMP12, and TOP2A expression in LUAD can lead to tumor development. It was also shown that these genes are closely associated with a poorer prognosis. Therefore, CRABP2, MMP12, and TOP2A may be useful biomarkers of LUAD and become key targets for its treatment. Further investigation is required however, to clarify the mechanisms involved in LUAD and comprehensively examine the role of hub genes in this context.
Three hub genes related to LUAD were identified via bioinformatics analysis: CRABP2, MMP12, and TOP2A. Increased expression of these genes can lead to the occurrence of LUAD, and is the most unfavorable prognostic factor in patients with LUAD. Hence, CRABP2, MMP12, and TOP2A may be important biomarkers of LUAD. To date limited research has been conducted on the roles of CRABP2, MMP12, and TOP2A in LUAD. The results of the current study provide a powerful impetus for investigating the pathogenesis of LUAD and developing favorable therapeutic targets in the future.
Adenocarcinoma of the lung (LUAD) is currently a cancer with high mortality. This study identified the biomarkers and therapeutic targets related to LUAD through bioinformatics analysis.
As of now, there are few biological analyses related to LUAD. Therefore, this study hopes to further study through big data analysis.
To determine reliable prognostic biomarkers for early diagnosis and treatment of Adenocarcinoma of the lung.
This article adopts bioinformatics methods such as gene Expression Omnibus (GEO) database, weighted gene co-expression network analysis, GEO2R, Gene Ontology analysis, protein-protein interaction network construction, University of Alabama at Birmingham Cancer data analysis portal database, etc.
We found three genes, namely, cellular retinoic acid binding protein 2, matrix Metalloprotein peptidase 12 and DNA Topoisomerase II α. The high expression of these genes is related to the poor prognosis of LUAD, and these Gene expression have high diagnostic value.
We identified genes related to LUAD treatment and prognosis through bioinformatics methods, providing important information for the complete cure of LUAD.
At present, there is little bioinformatics research related to LUAD. Through Big data screening, this study has more accurately identified the biomarkers and therapeutic targets related to LUAD, providing important information for the complete cure of LUAD in the future.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Mathematical and computational biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hashimoto K, Japan; Teixeira KN, Brazil S-Editor: Chen YL L-Editor: A P-Editor: Cai YX
| 1. | Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med. 2020;41:1-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 1162] [Article Influence: 232.4] [Reference Citation Analysis (0)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55853] [Article Influence: 7979.0] [Reference Citation Analysis (132)] |
| 3. | Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, Geisinger K, Hirsch FR, Ishikawa Y, Kerr KM, Noguchi M, Pelosi G, Powell CA, Tsao MS, Wistuba I; WHO Panel. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol. 2015;10:1243-1260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2160] [Cited by in RCA: 3130] [Article Influence: 347.8] [Reference Citation Analysis (0)] |
| 4. | Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584-594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 571] [Cited by in RCA: 1566] [Article Influence: 92.1] [Reference Citation Analysis (0)] |
| 5. | Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1023] [Cited by in RCA: 1399] [Article Influence: 127.2] [Reference Citation Analysis (0)] |
| 6. | Martin P, Leighl NB. Review of the use of pretest probability for molecular testing in non-small cell lung cancer and overview of new mutations that may affect clinical practice. Ther Adv Med Oncol. 2017;9:405-414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 7. | Goodgame B, Viswanathan A, Miller CR, Gao F, Meyers B, Battafarano RJ, Patterson A, Cooper J, Guthrie TJ, Bradley J, Pillot G, Govindan R. A clinical model to estimate recurrence risk in resected stage I non-small cell lung cancer. Am J Clin Oncol. 2008;31:22-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Fedrigo O, Naylor G. A gene-specific DNA sequencing chip for exploring molecular evolutionary change. Nucleic Acids Res. 2004;32:1208-1213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 9. | Cowell JK, Hawthorn L. The application of microarray technology to the analysis of the cancer genome. Curr Mol Med. 2007;7:103-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Schmidt U, Begley CG. Cancer diagnosis and microarrays. Int J Biochem Cell Biol. 2003;35:119-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Xu JY, Zhang C, Wang X, Zhai L, Ma Y, Mao Y, Qian K, Sun C, Liu Z, Jiang S, Wang M, Feng L, Zhao L, Liu P, Wang B, Zhao X, Xie H, Yang X, Chang Y, Jia J, Zhang Y, Wang Y, Yang Y, Wu Z, Yang L, Liu B, Zhao T, Ren S, Sun A, Zhao Y, Ying W, Wang F, Wang G, Cheng S, Qin J, Qian X, Li J, He F, Xiao T, Tan M. Integrative Proteomic Characterization of Human Lung Adenocarcinoma. Cell. 2020;182:245-261.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 363] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 12. | Wu X, Sui Z, Zhang H, Wang Y, Yu Z. Integrated Analysis of lncRNA-Mediated ceRNA Network in Lung Adenocarcinoma. Front Oncol. 2020;10:554759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 137] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 13. | Wang Y, Zhou Z, Chen L, Li Y, Chu X. Identification of key genes and biological pathways in lung adenocarcinoma via bioinformatics analysis. Mol Cell Biochem. 2021;476:931-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 14. | Tao Z, Shi A, Li R, Wang Y, Wang X, Zhao J. Microarray bioinformatics in cancer- a review. J BUON. 2017;22:838-843. [PubMed] |
| 15. | Li J, Miao B, Wang S, Dong W, Xu H, Si C, Wang W, Duan S, Lou J, Bao Z, Zeng H, Yang Z, Cheng W, Zhao F, Zeng J, Liu XS, Wu R, Shen Y, Chen Z, Chen S, Wang M; Hiplot Consortium. Hiplot: a comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Brief Bioinform. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 162] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 16. | Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10254] [Cited by in RCA: 16478] [Article Influence: 969.3] [Reference Citation Analysis (0)] |
| 17. | Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216-W221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2064] [Cited by in RCA: 3239] [Article Influence: 1079.7] [Reference Citation Analysis (0)] |
| 18. | Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2365] [Cited by in RCA: 4229] [Article Influence: 528.6] [Reference Citation Analysis (0)] |
| 19. | Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1613] [Cited by in RCA: 2327] [Article Influence: 290.9] [Reference Citation Analysis (0)] |
| 20. | Xu F, Huang X, Li Y, Chen Y, Lin L. m(6)A-related lncRNAs are potential biomarkers for predicting prognoses and immune responses in patients with LUAD. Mol Ther Nucleic Acids. 2021;24:780-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 150] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 21. | Islam KM, Jiang X, Anggondowati T, Lin G, Ganti AK. Comorbidity and Survival in Lung Cancer Patients. Cancer Epidemiol Biomarkers Prev. 2015;24:1079-1085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 140] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 22. | Zhang D, Jiang Q, Ge X, Shi Y, Ye T, Mi Y, Xie T, Li Q, Ye Q. RHOV promotes lung adenocarcinoma cell growth and metastasis through JNK/c-Jun pathway. Int J Biol Sci. 2021;17:2622-2632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 23. | Lu T, Yang X, Huang Y, Zhao M, Li M, Ma K, Yin J, Zhan C, Wang Q. Trends in the incidence, treatment, and survival of patients with lung cancer in the last four decades. Cancer Manag Res. 2019;11:943-953. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 373] [Article Influence: 62.2] [Reference Citation Analysis (0)] |
| 24. | Buzdin A, Tkachev V, Zolotovskaia M, Garazha A, Moshkovskii S, Borisov N, Gaifullin N, Sorokin M, Suntsova M. Using proteomic and transcriptomic data to assess activation of intracellular molecular pathways. Adv Protein Chem Struct Biol. 2021;127:1-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Yang R, Zhou Y, Du C, Wu Y. Bioinformatics analysis of differentially expressed genes in tumor and paracancerous tissues of patients with lung adenocarcinoma. J Thorac Dis. 2020;12:7355-7364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | Liu Z, Sun D, Zhu Q, Liu X. The screening of immune-related biomarkers for prognosis of lung adenocarcinoma. Bioengineered. 2021;12:1273-1285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |