Published online Dec 26, 2014. doi: 10.5662/wjm.v4.i4.219
Revised: September 11, 2014
Accepted: October 14, 2014
Published online: December 26, 2014
Processing time: 386 Days and 10.8 Hours
AIM: To test the methodical and pre-analytical performance of a new multiplex cancer biomarker panel using magnetic beads.
METHODS: The MILLIPLEX® MAP Human Circulating Cancer Biomarker Magnetic Bead Panel 1 comprises the tumor markers carcinoembryonic antigen, alpha-fetoprotein, total prostate-specific antigen, cancer antigen 15-3, cancer antigen 19-9, cancer antigen 125, cytokeratine 19-fragment, β-human chorionic gonadotropin, human epididymis protein 4, osteopontin, prolactin, the cell death and angiogenesis markers soluble Fas, soluble Fas-ligand, tumor necrosis factor related apoptosis-inducing ligand, vascular endothelial growth factor and the immunological markers interleukin-6 (IL-6), IL-8, tumor necrosis factor-α, transforming growth factor α, fibroblast growth factor-2, macrophage migration inhibitory factor, leptin, hepatocyte growth factor, and stem cell factor. We determined intra- and inter-assay imprecision as well as dilution linearity using quality controls and serum pools. Furthermore, the stability of the 24 biomarkers examined in this panel was ascertained by testing the influence of different storage temperatures and time span before centrifugation.
RESULTS: For all markers measured in the synthetic internal quality controls, the intra-assay imprecision ranged between 2.26% and 9.41%, while for 20 of 24 measured markers in the physiological serum pools, it ranged between 1.68% and 12.87%. The inter-assay imprecision ranged between 1.48%-17.12% for 23 biomarkers in synthetic, and between 4.59%-23.88% for 18 biomarkers in physiological quality controls. Here, single markers with very low concentration levels had increased imprecision rates. Dilution linearity was acceptable (70%-130% recovery) for 20 biomarkers. Regarding pre-analytical influencing factors, most markers were stable if blood centrifugation was delayed or if serum was stored for up to 24 h at 4 °C and 25 °C after centrifugation. Comparable results were obtained in serum and plasma for most markers. However, great changes were observed for single markers.
CONCLUSION: MILLIPLEX® MAP Human Circulating Cancer Biomarker Magnetic Bead Panel 1 assay is a stable and precise method for detection of most biomarkers included in the kit. However, single markers have to be interpreted with care.
Core tip: In this study, the methodological quality of a new research-use-only multiplex magnetic bead assay, particularly designed for cancer diagnosis, was evaluated. This attractive panel includes 24 biomarkers: established as well as auspicious tumor markers and markers deriving from the fields of apoptosis, immunology and angiogenesis. Herewith, the complexity and multifactorial background of a cancer disease is depicted. Measurements were performed with physiological serum pools and intra- and inter-assay imprecision as well as dilution linearity were assessed. Furthermore, the influence of preanalytical factors was investigated.
- Citation: Hermann N, Dreßen K, Schildberg FA, Jakobs C, Holdenrieder S. Methodical and pre-analytical characteristics of a multiplex cancer biomarker immunoassay. World J Methodol 2014; 4(4): 219-231
- URL: https://www.wjgnet.com/2222-0682/full/v4/i4/219.htm
- DOI: https://dx.doi.org/10.5662/wjm.v4.i4.219
Despite of essential achievements in cancer research concerning diagnosis, therapy options and follow up methods, cancer diseases still present a global health problem[1]. A great variety of clinical and imaging tools are applied to diagnose tumor masses and screening programs have been established for certain entities[2]. Some serum tumor markers, such as alpha-fetoprotein (AFP), cancer antigen 125 (CA 125), CA 15-3, CA 19-9, carcinoembryonic antigen (CEA) or prostate-specific antigen (PSA), have been introduced as supplementary diagnostic tools, but none of the above is recommended as a singular method to define a cancer diagnosis[3-5].
Cancer is nowadays perceived as a complex disease involving inflammatory and immunological systems and programs of cell death[6,7]. Thus, the diagnostic opportunity could be greatly enhanced by measurement of more than one marker as a fraction of information required to understand a complex pathological state[8,9]. Based on these findings, methods for parallel tumor marker testing have become more and more interesting in cancer research. Here, biomarkers, representing different systemic processes, such as inflammation, angiogenesis or cell death, can be combined with established tumor markers in one panel and potentially increase diagnostic accuracy[10-12].
Multiplex based immunoassays belong to the leading methods in this field. They are based on flow cytometry principles applied to labeled microspheres and depict an “ELISA on a bead”[13]. They offer several advantages, such as high-throughput performance, low material requirement, wide range application and cost- and time-effective multiplexing of more than 20 parameters[8,13].
However, the implementation of bead based multiplex assays has not yet been established in clinical routine[14]. Currently used tumor markers are mainly tested with single parameter assays. Not least due to the great potential of differently composed assays or marker panels, respectively, this field requires further research in order to assess assay quality, increase comparability of multiplex assays, and to encourage consistent guidelines which as of yet are non-existent[3,9,15].
As already shown by other research groups marker combination has the potential to greatly improve the quality of early diagnosis and other therapeutically relevant applications[12,16,17]. Several manufacturers offer diverse panels of markers, mainly for the combined measurement of many immunological and metabolic markers. For oncological purposes, the MILLIPLEX® Map Human Circulating Cancer Biomarker Magnetic Bead Panel Kit (EMD Millipore) was recently released. It represents an attractive option particularly for study settings. This kit includes reagents for the detection of 24 biomarkers, which portray a widespread spectrum of already validated as well as upcoming auspicious oncological, cell death, angiogenesis and immunological biomarkers, such as CEA, AFP, total prostate-specific antigen (total-PSA), CA 15-3, CA 19-9, CA 125, cytokeratine 19-fragment (CYFRA 21-1), β-human chorionic gonadotropin (β-HCG), human epididymis protein 4 (HE4), osteopontin (OPN), prolactin, soluble Fas (sFas), soluble Fas-ligand (sFasL), tumor necrosis factor related apoptosis-inducing ligand (TRAIL), vascular endothelial growth factor (VEGF), interleukin-6 (IL-6), IL-8, tumor necrosis factor-α (TNFα), transforming growth factor α (TGFα), fibroblast growth factor-2 (FGF2), macrophage migration inhibitory factor (MIF), leptin, hepatocyte growth factor (HGF) and stem cell factor (SCF).
However, in order to be used in studies and for clinical measurements, this panel must fulfill certain requirements, such as high reliability, accuracy, robustness as well as high analytical and clinical sensitivity and specificity[18,19]. Furthermore, the analytes must be stable against potentially influencing pre-analytical factors[3]. This study was carried out to critically test whether all or only some of the markers fulfill these basic methodical quality criteria and can thus be recommended for application in clinical or study conditions.
In order to assess the methodological performance of MILLIPLEX® MAP Human Circulating Cancer Biomarker Magnetic Bead Panel 1, 96 Well Plate Assay we tested intra- and inter-assay imprecision as well as dilution linearity.
Standard samples plus quality controls QC 1 and QC 2 delivered by the kits were used for the internal methodological control. Standard 7 depicted the basis for a dilution line with the factor 1:3 from high to lower biomarker concentrations. The standard dilution line as well as the concentrations of QC 1 and QC 2 were predefined by the manufacturer.
For external control, we produced two serum pools with levels in the moderate to high and in the very low value range for most markers (pool 1 and pool 2). To create pool 1, 37 residual and anonymized sera of daily clinical routine diagnostics were combined. Inclusion criteria were present high levels of the inflammation parameter C-reactive protein and well above average levels of the biomarkers AFP, β-HCG, CA 15-3, CA 125, CA 19-9, CEA, NSE and PSA. Here, patient history was not considered. Pool 2 is a combination of two sera taken from young healthy women (mean age 23.5 years). The sera pools, standard samples and quality controls QC 1 and QC 2 were run in duplicate as minimum within each plate.
In order to evaluate the linearity of dilution, a 50% dilution of the higher concentrated pool 1 was prepared by mixing the pool 1 sample with the appropriate amount of serum matrix enclosed in our kit. We defined the acceptable range for the recoveries as values between 70% and 130%.
Next, the estimation of possible affecting pre-analytical issues was tested. Briefly, samples of two different patients were stored at 25 °C (room temperature) for 6 and 24 h prior to centrifugation and subsequent freezing at -80 °C. In a further experiment, samples were stored at 4 °C and 25 °C for 6, 24 and 48 h, respectively, after centrifugation and before freezing at -80 °C. As reference control we used the corresponding samples, which were directly frozen after centrifugation. Finally, biomarkers were tested in serum and EDTA-plasma samples that were taken in parallel from the two healthy donors. All different conditions were measured in a single plate at a later time point to avoid inter-assay interferences.
The MILLIPLEX® MAP Human Circulating Cancer Biomarker Magnetic Bead Panel 1, 96 well plate assay purchased from EMD Millipore included all the reagents as well as an appropriate plate required by the assay. The procedure was conducted by experienced staff according to the manufacturer’s protocol. For washing steps, the Bio-Plex® Pro II wash station was applied. All plates were run on the Bio-Plex® 200 System. Before each assay run, the system was calibrated with the Bio-Plex® calibration kit and validated with the Bio-Plex® validation kit 4.0. Bio-Plex® sheath fluid served as the delivery medium for the samples. Analysis was performed with Bio-Plex® manager 6.1. Within the device settings, 50 events per bead region were defined as minimum criterion.
MILLIPLEX® MAP Kit Human Circulating Cancer Biomarker Magnetic Bead Panel 1 was developed as an immunoassay on the surface of fluorescent-coded magnetic beads (MagPlex™-C microspheres). The proportion of two fluorescent dyes on these beads forms the code and determines in such way up to 100 different kinds of beads. Here, we have 24 differently coded bead groups, each of which is coated with a specific capture antibody to detect one of the 24 biomarkers which are CEA, AFP, PSA, CA 15-3, CA 19-9, CA 125, CYFRA 21-1, β-HCG, HE4, osteopontin and prolactin, the cell death and angiogenesis markers sFas, sFasL, TRAIL and VEGF as well as the immunological markers IL-6, IL-8, TNFα, TGFα, FGF-2, MIF, leptin, HGF and SCF.
The binding of specific analytes begins in the bead mixture suspended with a test sample. Next, a biotinylated detection antibody is introduced and subsequent incubation with streptavidin-phycoerythrin (PE) conjugate is performed to complete the reaction on the microspheres.
Finally, the assay is analyzed by the Bio-Plex® 200 system. Here, the beads coupled with the capture antibody bound to the specific analyte, biotinylated detection antibody and streptavidin-PE on its surfaces pass through a laser, which excites the internal dyes. A second laser excites the signal of PE. High-speed digital-signal processors identify the beads and detect the fluorescent signal intensity in order to quantify the assay result.
All reagents and sera were brought to room temperature before use. Wash buffer, assay buffer, serum matrix, standard 7, quality controls 1 and 2, beads, detection antibodies, and streptavidin-PE were prepared as recommended by the manufacturer. Serum samples were thawed and individually vortexed for 15 s. Thereafter, they were centrifuged (Eppendorf centrifuge 581OR) at 3500 rpm for one min. Next, 15 μL of sample was mixed with 75 μL of serum matrix creating a 1:6 dilution. To create a homogeneous mixture of all antibody conjugated beads, every vial containing one set of microspheres was sonicated for 30 s and then vortexed for one minute. One hundred and fifty microliter of each vial was transferred into a mixing bottle that was vortexed again for one minute. The beads were protected from exposure to light throughout the assay. For pre-wetting, 200 μL assay buffer was pipetted into each well, the plate was covered with a sealer and then shaken at 700 rpm for 10 min. The fluid was removed by tapping the plate on a paper towel and centrifuging it briefly at 3500 rpm lying top down on a paper sheet in the centrifuge. Twenty-five microliter of background, standard 1-7 and quality controls 1 and 2 were pipetted in duplicate into the appropriate wells and 25 μL of serum matrix was added. Next, sample wells were filled with 25 μL of assay buffer and 25 μL of the diluted sera was pipetted into the appropriate wells. Finally, the magnetic bead mixture was vortexed for 1 min and 25 μL were pipetted into each well. The plate was sealed, covered with aluminum foil and then shaken at 700 rpm for 16 h at 4 °C. After incubation time the magnetic bead plates were washed as recommended by the method protocol three times using assay buffer by means of Bio-Plex® Pro II wash station. Then 25 μL of detection antibodies were added to each well, the plate was sealed and covered with aluminum foil and shaken at 700 rpm for 1 h. Now 25 μL of streptavidin-PE per well were added and the plate was again sealed, covered with aluminum foil and shaken at 700 rpm for 30 min. Thereon the three washing steps were performed as described above. One hundred microliter of sheath fluid were pipetted into each well, the plate sealed and covered with aluminum foil and shaken at 700 rpm for 5 min in order to resuspend the beads. Lastly, the plates were run on the Bio-Plex® 200 system.
Expected concentrations of each tested biomarker for standards 1-7 were entered into the system prior to running the assay. The device detects appropriate fluorescence intensities (FI) and creates a standard curve for each marker. These curves are generated by linking the measured FI values with the expected concentration of markers in standards 1-7. Further translation of FI-values in concentration levels of all the following samples is based on these curves. Depending on the five parameter logistic, the function possesses predefined points of accepted extrapolation, which are the minimum and maximum asymptotes. For the lower limits in our study, we accepted an extrapolation in round terms in the middle between the lowest standard and the minimum asymptote point. Due to the phenomenon of heteroscedasticity towards higher concentrations, here, the accepted extrapolation was defined as an approximation of the highest standard value. These limits consequently illustrate our measuring range.
As all physiological serum samples were diluted 1:6 with serum matrix included into the kit, the dilution factor 6 was considered by the software before yielding the final concentration of the samples. For convenience of comparability in added tables, we multiplied all non diluted concentrations (accepted measuring range, observed concentration of QC 1, QC 2 and standard 5) with the factor 6.
The results from the software of Bio-Plex® 200 contain the measured FI and when duplicates were run, also the corresponding means, standard deviation and coefficients of variation (CV in %) as well as the corresponding concentrations.
In order to assess intra- and inter-assay imprecision FI-based CVs were used. We also determined the CVs based on the observed concentration for the analysis. Means and ranges were calculated for all comparisons. In order to quantify the dilution linearity, we determined observed concentration-based recoveries related to the corresponding expected values for the 50% dilution of pool 1. The evaluation of the pre-analytical influence of different storage conditions is represented by calculated recoveries based on FI results.
All in all, we tested ten kits or rather ten plates. The first five plates were ordered as a batch and were measured subsequently and strictly under the same conditions. The following five plates were of the same lot, but were ordered and measured about six months later under different conditions. Therefore, the main method evaluation in our study is based on the first five kits. However, results of the overall evaluation are also shown. In one assay, a pipetting error of the pool samples occurred and the respective values were omitted.
The intra-assay imprecision as the mean CV (in %) over all plates for synthetic quality controls QC 1, QC 2 and standard 5 as well as for physiological serum pools 1 and 2 was calculated for five different magnetic plates relating to each tested biomarker. Here, only results within the measuring range were included.
In QC 1 and QC 2, 22 and 24 markers had an FI-based CV below 10%. In QC 2, 21 biomarkers were measured with a CV less than 5%. The CVs ranged between 3.81% (AFP) and 13.38% (FGF2) for QC 1 and between 2.06% (total PSA) and 5.56% (β-HCG) for QC 2. Observed concentration-based CVs ranged between 4.23% (AFP) and 9.41% (FGF2) for QC 1 and between 2.26% (TRAIL) and 7.69% (CA 19-9) for QC 2 (Table 1).
| QC 1 | QC 2 | St 5 | Pool 1 | Pool 2 | |||||||||||||
| Biomarker | Measuring | Unit | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc |
| range | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | ||
| CEA | 100-120000 | pg/mL | 2505 | 7.24 | 8.87 | 10933 | 2.32 | 3.39 | 2150 | 1.29 | 38897 | 2.29 | 3.98 | 466 | 6.77 | 7.78 | |
| AFP | 500-600000 | pg/mL | 11980 | 3.81 | 4.23 | 54865 | 2.4 | 3.29 | 10703 | 2.88 | 455084 | 3.46 | 12.87 | 1598 | 4.99 | 11.99 | |
| Total PSA | 50-60000 | pg/mL | 1230 | 5.97 | 7.9 | 5880 | 2.06 | 3.41 | 1057 | 1.92 | 7762 | 2.46 | 4.16 | < MR | < MR | < MR | |
| CA 15-3 | 0.5-600 | U/mL | 12 | 9.1 | 8.43 | 57 | 4.06 | 4.5 | 11 | 3.05 | 29 | 6.48 | 6.35 | 24 | 3.87 | 3.7 | |
| CA 19-9 | 2-3000 | U/mL | 61 | 9.54 | 7.35 | 292 | 5.27 | 7.69 | 54 | 3.2 | 33 | 3.35 | 2.66 | 10 | 1.65 | 1.75 | |
| CA 125 | 2-4000 | U/mL | 71 | 6.77 | 6.68 | 347 | 4.14 | 4.4 | 68 | 2.27 | 302 | 4.15 | 4.36 | 5.53 | 6.23 | 11.25 | |
| β-HCG | 0.2-400 | mU/mL | 8 | 8.86 | 5.54 | 39 | 5.56 | 4.25 | 7 | 1.54 | 0.65 | 6.37 | 36.09 | < MR | < MR | < MR | |
| CYFRA 21-1 | 500-900000 | pg/mL | 29454 | 7.92 | 6.98 | 155151 | 3.7 | 3.18 | 16614 | 1.52 | 9724 | 4.22 | 4.77 | 3506 | 8.07 | 13.12 | |
| HE4 | 2000-3000000 | pg/mL | 55648 | 8.73 | 6.98 | 291745 | 5.17 | 5.23 | 55123 | 4.1 | 2276 | 8.16 | 9.58 | < MR | < MR | < MR | |
| Prolactin | 500-600000 | pg/mL | 11868 | 5.69 | 4.73 | 60195 | 3.04 | 3.55 | 10922 | 2.61 | 12057 | 4.37 | 3.69 | 10765 | 3.08 | 2.52 | |
| Leptin | 500-600000 | pg/mL | 13056 | 8.97 | 5.05 | 62955 | 2.75 | 2.38 | 10971 | 1.94 | 13430 | 7.17 | 4.04 | 8468 | 2.78 | 1.48 | |
| OPN | 2000-3000000 | pg/mL | 53895 | 8.94 | 7.32 | 236975 | 3.72 | 6.02 | 43171 | 0.91 | 73954 | 4.46 | 4.01 | 10870 | 2.75 | 2.25 | |
| HGF | 100-120000 | pg/mL | 2452 | 9.76 | 8.64 | 12766 | 4.06 | 4.28 | 2154 | 2.86 | 785 | 3.11 | 2.86 | 283 | 5.65 | 7.44 | |
| MIF | 100-120000 | pg/mL | 2230 | 7.83 | 5.97 | 11061 | 4.5 | 4.45 | 2209 | 3.06 | 574 | 1.89 | 1.68 | 513 | 14.31 | 15.66 | |
| sFas | 100-150000 | pg/mL | 2486 | 6.3 | 5.53 | 12705 | 3.19 | 2.77 | 2779 | 2.4 | 3548 | 4.52 | 3.89 | 2217 | 1.67 | 1.47 | |
| sFasL | 50-60000 | pg/mL | 1219 | 5.65 | 5.71 | 5898 | 2.17 | 2.28 | 1091 | 2.86 | < MR | < MR | < MR | < MR | < MR | < MR | |
| TRAIL | 10-12000 | pg/mL | 252 | 7.23 | 7.41 | 1233 | 2.16 | 2.26 | 221 | 2.49 | 65 | 3.4 | 3.62 | 92 | 3.32 | 3.5 | |
| VEGF | 50-60000 | pg/mL | 1127 | 13.04 | 8.8 | 6110 | 3.97 | 2.78 | 1115 | 4.41 | < MR | < MR | < MR | < MR | < MR | < MR | |
| IL-6 | 2-3000 | pg/mL | 49 | 9.2 | 9.27 | 226 | 4.33 | 4.21 | 53 | 3.43 | 16 | 5.66 | 7.02 | < MR | < MR | < MR | |
| IL-8 | 5-6000 | pg/mL | 122 | 5.53 | 5.67 | 605 | 2.19 | 2.34 | 110 | 1.71 | 33 | 5.85 | 6.7 | 6.71 | 4.49 | 9.43 | |
| TNFα | 5-6000 | pg/mL | 114 | 7.77 | 7.97 | 562 | 2.88 | 3.17 | 110 | 2.25 | 13 | 4.56 | 5.66 | 6.41 | 3.12 | 6.68 | |
| TGFα | 10-12000 | pg/mL | 237 | 6.36 | 6.29 | 1180 | 3.02 | 3.11 | 221 | 1.23 | 24 | 5.17 | 8.75 | 18 | 4.49 | 11.59 | |
| FGF2 | 50-50000 | pg/mL | 1101 | 13.38 | 9.41 | 5571 | 2.61 | 2.85 | 1066 | 3.08 | 55 | 8.71 | 18.11 | 79 | 9.57 | 14.52 | |
| SCF | 20-30000 | pg/mL | 599 | 8.41 | 7.97 | 2813 | 3.02 | 2.88 | 550 | 1.17 | 58 | 6.99 | 10.97 | 50 | 8.32 | 13.4 | |
In the standard 5-sample, all of the 24 biomarkers were measured with CV values below 5%. The range was between 0.91% (OPN)-4.41% (VEGF).
In the physiological serum pool 1, all biomarkers showed an FI-based CV below 10%, while 13 biomarkers had a CV below 5%. The values ranged from 1.89% (MIF) to 8.71% (FGF2). Observed concentration-based CVs ranged from 1.68% (MIF) to 36.09% (β-HCG) with 12 biomarkers measured with a CV below 5% and four biomarkers exceeding the 10% range (Table 1).
In physiological serum pool 2, FI-based imprecision ranged from 1.65% (CA 19-9) to 14.31% (MIF) with only one CV (MIF) found to be higher than 10%. CVs of 11 biomarkers fell below 5%. Observed concentration-based imprecision ranged from 1.47% (sFas)-15.66% (MIF) with seven biomarkers measured with a CV below 5% and seven biomarkers exceeding the 10% range (Table 1).
When intra-assay imprecision was evaluated for all ten plates, the CVs were somewhat higher for pools and QC samples (Table 2).
| QC 1 | QC 2 | St 5 | Pool 1 | Pool 2 | |||||||||||||
| Biomarker | Measuring | Unit | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc |
| range | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | ||
| CEA | 100-120000 | pg/mL | 2545 | 8.32 | 11.34 | 12727 | 7.61 | 12.23 | 2146 | 4.11 | 39391 | 2.1 | 3.63 | 560 | 8.09 | 5.67 | |
| AFP | 500-600000 | pg/mL | 12038 | 6.09 | 6.9 | 56960 | 3.08 | 4.23 | 10754 | 4.45 | 493993 | 2.39 | 8.73 | 1489 | 6.87 | 11.8 | |
| Total PSA | 50-60000 | pg/mL | 1421 | 5.76 | 7.78 | 6172 | 3.75 | 6.27 | 1052 | 4.1 | 8246 | 1.59 | 2.69 | 68 | 9.76 | 7.02 | |
| CA 15-3 | 0.5-600 | U/mL | 12 | 9 | 8.7 | 62 | 4.18 | 4.96 | 11 | 4.32 | 33 | 5.31 | 5.23 | 26 | 9.19 | 8.66 | |
| CA 19-9 | 2-3000 | U/mL | 64 | 10.63 | 8.63 | 329 | 4.83 | 7.1 | 54 | 5.05 | 36 | 2.9 | 2.39 | 10 | 4.87 | 6.17 | |
| CA 125 | 2-4000 | U/mL | 74 | 7.02 | 7.03 | 370 | 3.61 | 3.78 | 68 | 2.84 | 317 | 3.59 | 3.64 | 6.13 | 4.74 | 8.54 | |
| β-HCG | 0.2-400 | mU/mL | 7.85 | 10.62 | 7.02 | 42 | 4.89 | 3.67 | 7 | 5.08 | 1.2 | 6.76 | 18.28 | 0.55 | 8.17 | 31.39 | |
| CYFRA 21-1 | 500-900000 | pg/mL | 24985 | 7.62 | 8.22 | 152731 | 12.13 | 4.28 | 16601 | 3.32 | 8627 | 4.34 | 5.73 | 3506 | 8.33 | 13.12 | |
| HE4 | 2000-3000000 | pg/mL | 55766 | 11.42 | 9.39 | 295050 | 4.28 | 4.45 | 55043 | 1.9 | 2288 | 6.47 | 7.67 | 2590 | 11.67 | 8.86 | |
| Prolactin | 500-600000 | pg/mL | 11952 | 7.86 | 6.96 | 60823 | 3.43 | 4.13 | 10863 | 4.49 | 12191 | 3.31 | 2.83 | 10602 | 6.1 | 4.71 | |
| Leptin | 500-600000 | pg/mL | 13001 | 12.79 | 7.06 | 64533 | 2.79 | 2.4 | 11020 | 5.73 | 14622 | 5.67 | 3.18 | 9145 | 6.58 | 3.4 | |
| OPN | 2000-3000000 | pg/mL | 57576 | 9.61 | 8.58 | 240688 | 4.93 | 9.29 | 42845 | 3.77 | 75488 | 3.61 | 3.35 | 10700 | 5.59 | 4.85 | |
| HGF | 100-120000 | pg/mL | 2546 | 9.92 | 8.68 | 13471 | 3.23 | 3.3 | 2171 | 2.51 | 894 | 2.82 | 2.53 | 436 | 7.6 | 8.8 | |
| MIF | 100-120000 | pg/mL | 2263 | 10.84 | 10.06 | 12332 | 5.12 | 5.99 | 2220 | 5.19 | 876 | 2.06 | 1.86 | 517 | 11.4 | 17.25 | |
| sFas | 100-150000 | pg/mL | 2533 | 7.15 | 6.72 | 13212 | 3.91 | 3.47 | 2775 | 3.94 | 4005 | 3.54 | 3.07 | 2472 | 4.88 | 4.34 | |
| sFasL | 50-60000 | pg/mL | 1245 | 6.35 | 6.49 | 6023 | 2.9 | 3.15 | 1093 | 5.13 | < MR | < MR | < MR | < MR | < MR | < MR | |
| TRAIL | 10-12000 | pg/mL | 253 | 8.52 | 8.64 | 1269 | 2.65 | 2.72 | 220 | 3.73 | 72 | 4.22 | 4.71 | 98 | 6.19 | 6.44 | |
| VEGF | 50-60000 | pg/mL | 1146 | 16.04 | 11.12 | 6155 | 4.61 | 3.56 | 1100 | 2.88 | < MR | < MR | < MR | <MR | < MR | < MR | |
| IL-6 | 2-3000 | pg/mL | 54 | 11.72 | 12.01 | 268 | 6.35 | 6.11 | 54 | 1.79 | 21 | 6.25 | 7.44 | < MR | < MR | < MR | |
| IL-8 | 5-6000 | pg/mL | 124 | 7.8 | 9.02 | 668 | 6.56 | 9.49 | 111 | 3.94 | 26 | 5.05 | 9.13 | 6.70 | 6.22 | 9.43 | |
| TNFα | 5-6000 | pg/mL | 117 | 10.43 | 10.35 | 583 | 3.68 | 3.8 | 111 | 4.86 | 16 | 5.27 | 9.14 | 7.85 | 5.41 | 4.61 | |
| TGFα | 10-12000 | pg/mL | 238 | 8.86 | 8.94 | 1210 | 3.78 | 4.05 | 221 | 4.91 | 27 | 3.99 | 9.09 | 18 | 5.32 | 9.69 | |
| FGF2 | 50-50000 | pg/mL | 1104 | 20.18 | 14.17 | 5708 | 2.97 | 3.07 | 1068 | 2.04 | 68 | 9.63 | 13.35 | 80 | 7.72 | 7.97 | |
| SCF | 20-30000 | pg/mL | 603 | 8.04 | 7.66 | 2951 | 4.39 | 4.32 | 549 | 3.96 | 79 | 6.49 | 8.93 | 59 | 8.5 | 15.12 | |
FI-based imprecision: The inter-assay imprecision was performed by calculating the CV in % involving all fluorescence intensity results for QC 1, QC 2, standard 5, pool 1 and pool 2 over five different magnetic plates relating to each tested marker. Here, only results within the measuring range were included.
For synthetic QC 1, QC 2 and standard 5, 16, 17 and 18 markers had imprecision below 20%, ranging between 8.85% (IL-8) and 45.75% (OPN), 8.88% (CEA) and 29.04% (OPN) as well as between 8.61% (total PSA) and 42.71% (CYFRA 21-1) (Table 3).
| QC 1 | QC 2 | St 5 | Pool 1 | Pool 2 | |||||||||||||
| Biomarker | Measuring | Unit | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc |
| range | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | ||
| CEA | 100-120000 | pg/mL | 2505 | 10.45 | 3.92 | 10933 | 8.88 | 7.62 | 2150 | 11.53 | 4.15 | 38897 | 4.53 | 6.58 | 466 | 15.14 | 14.43 |
| AFP | 500-600000 | pg/mL | 11980 | 13.92 | 5.32 | 54865 | 12.72 | 5.09 | 10703 | 12.14 | 3.43 | 455084 | 8.59 | 20.68 | 1598 | 34 | 66.04 |
| Total PSA | 50-60000 | pg/mL | 1230 | 15.82 | 5.11 | 5880 | 11.51 | 11.21 | 1057 | 8.61 | 0.32 | 7762 | 7.88 | 4.59 | < MR | < MR | < MR |
| CA 15-3 | 0.5-600 | U/mL | 12 | 18.57 | 6.38 | 57 | 21.29 | 2.85 | 11 | 20.52 | 1.79 | 29 | 16.48 | 8.48 | 24 | 27.78 | 21.2 |
| CA 19-9 | 2-3000 | U/mL | 61 | 20.3 | 4.79 | 292 | 12.19 | 4.58 | 54 | 11.44 | 2.19 | 33 | 17.49 | 7.29 | 10 | 19.24 | 23.19 |
| CA 125 | 2-4000 | U/mL | 71 | 13.39 | 3.23 | 347 | 11.19 | 3.9 | 68 | 11.73 | 1.91 | 302 | 6.26 | 8.27 | 5.53 | 22.25 | 36.3 |
| β-HCG | 0.2-400 | mU/mL | 8 | 17.8 | 4.39 | 39 | 16.3 | 1.48 | 7 | 15.48 | 2.41 | 0.65 | 4.49 | 61.53 | < MR | < MR | < MR |
| CYFRA 21-1 | 500-900000 | pg/mL | 29454 | 16.78 | 27.37 | 155151 | 15.64 | 40.96 | 16614 | 42.71 | 1.14 | 9724 | 38.02 | 50 | 3506 | 21.25 | 30.8 |
| HE4 | 2000-3000000 | pg/mL | 55648 | 27.52 | 5.48 | 291745 | 23.45 | 3.46 | 55123 | 23.76 | 2.74 | 2276 | 14.67 | 63.85 | < MR | < MR | < MR |
| Prolactin | 500-600000 | pg/mL | 11868 | 26.35 | 4.16 | 60195 | 15.98 | 2.69 | 10922 | 14.69 | 1.97 | 12057 | 21.22 | 10.08 | 10765 | 35.99 | 20.91 |
| Leptin | 500-600000 | pg/mL | 13056 | 23.59 | 2.44 | 62955 | 14.97 | 3.92 | 10971 | 15.63 | 1.81 | 13430 | 18.98 | 8.79 | 8468 | 36.1 | 23.77 |
| OPN | 2000-3000000 | pg/mL | 53895 | 45.75 | 4.01 | 236975 | 29.04 | 4.62 | 43171 | 31.41 | 2.38 | 73954 | 46.72 | 8.5 | 10870 | 85.54 | 22.7 |
| HGF | 100-120000 | pg/mL | 2452 | 13.41 | 12.19 | 12766 | 16.96 | 8.28 | 2154 | 20.58 | 1.7 | 785 | 7.96 | 11.57 | 283 | 10.19 | 20.14 |
| MIF | 100-120000 | pg/mL | 2230 | 19.69 | 7.89 | 11061 | 14.32 | 4.89 | 2209 | 12.31 | 1.68 | 574 | 8.59 | 13.68 | 513 | 63.56 | 62.9 |
| sFas | 100-150000 | pg/mL | 2486 | 27.52 | 6.36 | 12705 | 21.86 | 6.83 | 2779 | 26.23 | 1.37 | 3548 | 30.07 | 7.78 | 2217 | 51.45 | 22.99 |
| sFasL | 50-60000 | pg/mL | 1219 | 16.69 | 4.68 | 5898 | 20.4 | 2.92 | 1091 | 17.72 | 1.62 | < MR | < MR | < MR | < MR | < MR | < MR |
| TRAIL | 10-12000 | pg/mL | 252 | 13.53 | 4.72 | 1233 | 13.51 | 4.09 | 221 | 15.17 | 1.85 | 65 | 10.05 | 9.26 | 92 | 27.2 | 23.98 |
| VEGF | 50-60000 | pg/mL | 1127 | 25.4 | 4.27 | 6110 | 24.51 | 8.87 | 1115 | 18.99 | 3.76 | < MR | < MR | < MR | < MR | < MR | < MR |
| IL-6 | 2-3000 | pg/mL | 49 | 14.49 | 9.34 | 226 | 22.21 | 17.12 | 53 | 18.81 | 4 | 16 | 12.3 | 14.64 | < MR | < MR | < MR |
| IL-8 | 5-6000 | pg/mL | 122 | 8.85 | 5.21 | 605 | 11.32 | 3.73 | 110 | 9.31 | 1.98 | 33 | 9.31 | 8.37 | 6.71 | 25.35 | 40.54 |
| TNFα | 5-6000 | pg/mL | 114 | 21.44 | 5.52 | 562 | 16.31 | 6.81 | 110 | 16.78 | 2.28 | 13 | 19.54 | 8.14 | 6.41 | 26.96 | 40.75 |
| TGFα | 10-12000 | pg/mL | 237 | 16.15 | 6.26 | 1180 | 13.07 | 3.33 | 221 | 15.5 | 1.97 | 24 | 16.87 | 23.88 | 18 | 24.27 | 32.78 |
| FGF2 | 50-50000 | pg/mL | 1101 | 11.17 | 8.98 | 5571 | 13.86 | 5.71 | 1066 | 12.75 | 3.89 | 55 | 11.36 | 68.81 | 79 | 19.8 | 40.04 |
| SCF | 20-30000 | pg/mL | 599 | 15.96 | 10.08 | 2813 | 13.38 | 4.75 | 550 | 14.69 | 1.93 | 58 | 11.5 | 10.22 | 50 | 24.27 | 22.5 |
In the higher-concentrated physiological serum pool 1, the inter-assay imprecision fell below 20% for 18 biomarkers with the total range between 4.49% (β-HCG) and 46.72% (OPN). In the very low serum pool 2, only four biomarkers were measured with a CV below 20%, collectively ranging between 10.19% (HGF) and 85.54% (OPN) (Table 3).
Observed concentration based imprecision: In synthetic internal controls, the imprecision was below 20% for 23 biomarkers in QC 1 and QC 2. The same applied to 24 biomarkers in standard 5. The corresponding ranges were 2.44% (Leptin) to 27.37% (CYFRA 21-1), 1.48% (β-HCG) to 40.96% (CYFRA 21-1) and 0.32% (total PSA) to 4.15% (CEA) (Table 3).
In the physiological serum pool 1, the imprecision fell below 20% for 16 biomarkers and values ranged from 4.59% (total PSA) to 68.81% (FGF2). Notably, single markers in pool 1 with very low concentration levels had considerably higher imprecision rates. In serum pool 2 with very low values for all markers, only one marker (CEA) was measured with a CV less than 20%. Here, the imprecision ranged in total between 14.43% (CEA)-66.04% (AFP) (Table 3).
When inter-assay imprecision was evaluated for all ten plates, the CVs were somewhat higher for pools and QC samples (Table 4).
| QC 1 | QC 2 | St 5 | Pool 1 | Pool 2 | |||||||||||||
| Biomarker | Measuring | Unit | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc | Conc | FI | Conc |
| range | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | Mean | Mean CV% | Mean CV% | ||
| CEA | 100-120000 | pg/mL | 2545 | 13.96 | 28.08 | 12727 | 20.57 | 25.67 | 2146 | 13.34 | 4.59 | 39391 | 6.94 | 7.38 | 560 | 14.32 | 25.77 |
| AFP | 500-600000 | pg/mL | 12038 | 23.54 | 20.99 | 56960 | 19.11 | 10.29 | 10754 | 21.43 | 3.25 | 493993 | 12.38 | 18.42 | 1489 | 33.13 | 63.64 |
| Total PSA | 50-60000 | pg/mL | 1421 | 34.29 | 31.87 | 6172 | 17.44 | 13.78 | 1052 | 18.72 | 3.67 | 8246 | 16.03 | 12.46 | 68 | 130.09 | 151.66 |
| CA 15-3 | 0.5-600 | U/mL | 12 | 30.11 | 18.58 | 62 | 29.25 | 12.98 | 11 | 34.01 | 2.93 | 33 | 23.72 | 17.26 | 26 | 30.49 | 27.59 |
| CA 19-9 | 2-3000 | U/mL | 64 | 22.71 | 17.91 | 329 | 11.98 | 19.1 | 54 | 19.2 | 2.71 | 36 | 15.18 | 26.88 | 10 | 16.18 | 36.06 |
| CA 125 | 2-4000 | U/mL | 74 | 14.42 | 22.04 | 370 | 12.5 | 11.07 | 68 | 17.29 | 2.42 | 317 | 7.72 | 12.62 | 6.13 | 18.13 | 37.03 |
| β-HCG | 0.2-400 | mU/mL | 7.85 | 20.62 | 20.74 | 42 | 16.95 | 16.23 | 7 | 26.88 | 2.69 | 1.2 | 29.09 | 52.2 | 0.55 | 17.3 | 74.36 |
| CYFRA 21-1 | 500-900000 | pg/mL | 24985 | 69.07 | 36.07 | 152731 | 81.41 | 52.97 | 16601 | 53.61 | 2.5 | 8627 | 50.45 | 55.77 | 3506 | 31.82 | 30.8 |
| HE4 | 2000-3000000 | pg/mL | 55766 | 22.11 | 21.46 | 295050 | 19.92 | 10.63 | 55043 | 22.45 | 2.1 | 2288 | 19.7 | 68.01 | 2590 | 22.73 | 54.77 |
| Prolactin | 500-600000 | pg/mL | 11952 | 23.76 | 21.49 | 60823 | 13.52 | 9.33 | 10863 | 14.81 | 2.05 | 12191 | 17.16 | 20.41 | 10602 | 29.43 | 28.46 |
| Leptin | 500-600000 | pg/mL | 13001 | 29.19 | 15.47 | 64533 | 19.61 | 8.82 | 11020 | 23.3 | 1.88 | 14622 | 16.75 | 18.61 | 9145 | 31.06 | 27.03 |
| OPN | 2000-3000000 | pg/mL | 57576 | 58.11 | 28.1 | 240688 | 24.08 | 13.88 | 42845 | 25.18 | 5.71 | 75488 | 41.77 | 25.01 | 10700 | 62.83 | 47.91 |
| HGF | 100-120000 | pg/mL | 2546 | 15.19 | 20.7 | 13471 | 19.05 | 12.85 | 2171 | 22.65 | 2.04 | 894 | 7.18 | 27.83 | 436 | 29.6 | 49.81 |
| MIF | 100-120000 | pg/mL | 2263 | 46.52 | 45.1 | 12332 | 20.48 | 18.05 | 2220 | 23.01 | 2.93 | 876 | 62.88 | 48.48 | 517 | 45.1 | 47.35 |
| sFas | 100-150000 | pg/mL | 2533 | 27.15 | 18.62 | 13212 | 20.06 | 12.63 | 2775 | 27.38 | 1.77 | 4005 | 29.39 | 21.39 | 2472 | 39.44 | 28.26 |
| sFasL | 50-60000 | pg/mL | 1245 | 19.09 | 20.91 | 6023 | 21.37 | 6.63 | 1093 | 22.37 | 1.98 | < MR | < MR | < MR | < MR | < MR | < MR |
| TRAIL | 10-12000 | pg/mL | 253 | 20.17 | 21.78 | 1269 | 17.13 | 10.13 | 220 | 21.55 | 2.05 | 72 | 11.04 | 32.6 | 98 | 26.7 | 35.57 |
| VEGF | 50-60000 | pg/mL | 1146 | 46.21 | 23 | 6155 | 45.49 | 8.97 | 1100 | 43.93 | 3.26 | < MR | < MR | < MR | < MR | < MR | < MR |
| IL-6 | 2-3000 | pg/mL | 54 | 17.46 | 26.94 | 268 | 17.39 | 23.92 | 54 | 24.51 | 5.54 | 21 | 14.19 | 41.05 | < MR | < MR | < MR |
| IL-8 | 5-6000 | pg/mL | 124 | 75.72 | 21.22 | 668 | 45.79 | 22.2 | 111 | 30.7 | 3.13 | 26 | 36.91 | 38.62 | 6.70 | 90.89 | 40.54 |
| TNFα | 5-6000 | pg/mL | 117 | 18.6 | 23.55 | 583 | 16.6 | 11.16 | 111 | 19.41 | 2.52 | 16 | 15.5 | 28.1 | 7.85 | 22.35 | 49.24 |
| TGFα | 10-12000 | pg/mL | 238 | 18.16 | 21.43 | 1210 | 19.49 | 7.74 | 221 | 22.29 | 2.56 | 27 | 17.64 | 45.12 | 18 | 19.22 | 48.85 |
| FGF2 | 50-50000 | pg/mL | 1104 | 25.12 | 23.52 | 5708 | 17.67 | 9.9 | 1068 | 19.05 | 3.55 | 68 | 9.43 | 49.91 | 80 | 15.21 | 32.41 |
| SCF | 20-30000 | pg/mL | 603 | 12.44 | 19.52 | 2951 | 16.66 | 11.26 | 549 | 17.88 | 2.01 | 79 | 31.9 | 40.75 | 59 | 22.35 | 32.17 |
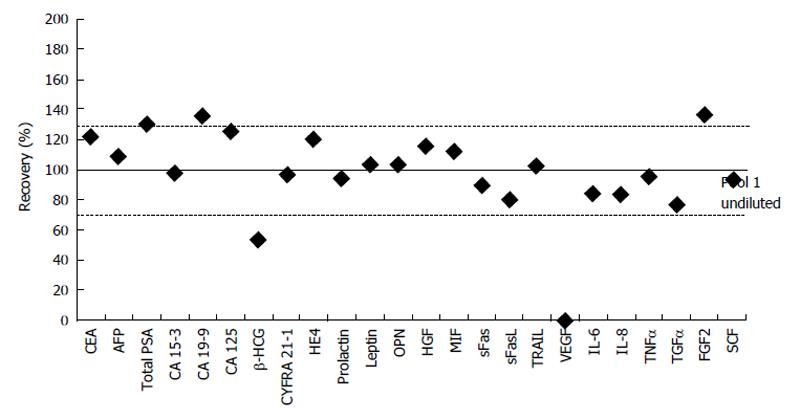
The 50% dilutions of pool 1 samples were run in each of the five plates. The dilution of 20 biomarkers fell into the accepted recovery of 70%-130%. Range of all markers was between 53.19% (β-HCG) and 136.24% (FGF2) (Figure 1). Here, concentration levels calculated by extrapolation were included. VEGF was the only biomarker without any calculable levels of concentration.
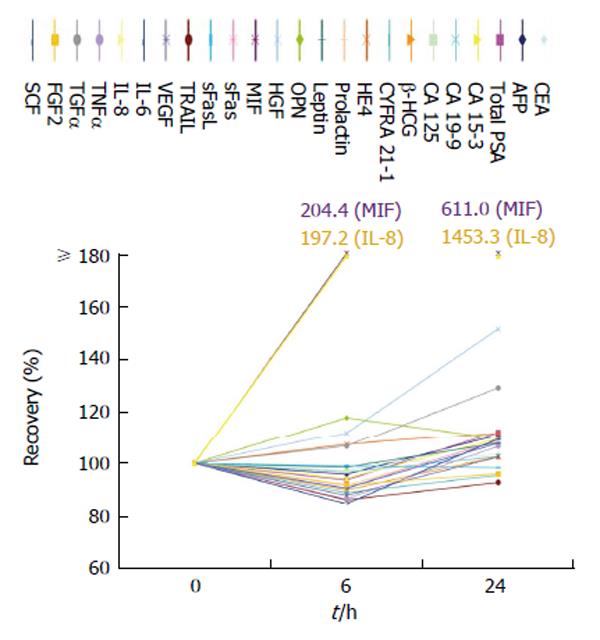
Samples centrifuged and measured after being stored for 6 h at room temperature (25 °C) yielded a median recovery of 95.0% [range: 84.5% (IL-6)-204.4% (MIF)], while a centrifugation after 24 h showed a stronger effect on some biomarkers with a median recovery of 108.2% ranging from 92.8% for TRAIL to 1453.3% for IL-8. However, only two biomarkers after 6 h and three biomarkers after 24 h failed the accepted range of recovery (IL-8 and MIF) (Figure 2).
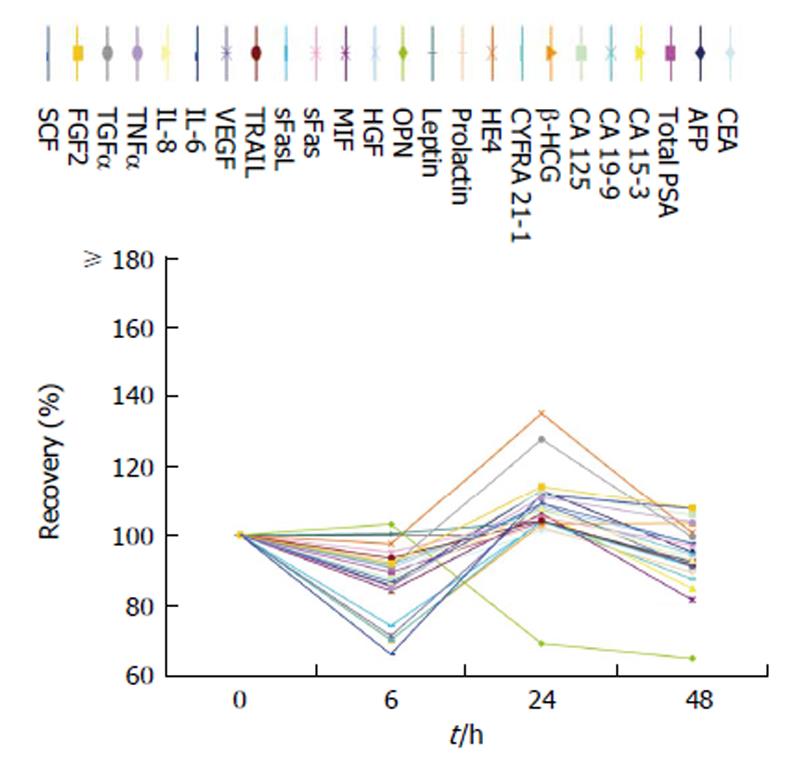
When samples were directly centrifuged after venous puncture and subsequently stored at 4 °C up to 48 h before measurements, biomarker levels showed relatively stable results. When storing the samples at room temperature for 6, 24 and 48 h before testing, stronger alterations were observed. Indeed, two, 0 and 16 biomarkers, respectively, failed the 70%-130% range of recovery. Detailed results are shown in Figures 3 and 4.
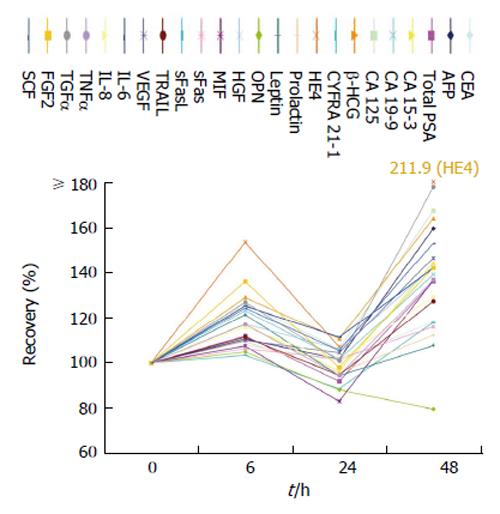
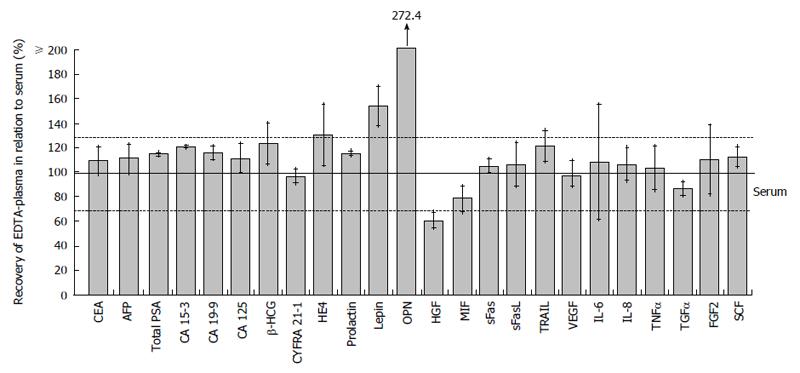
Testing in EDTA-plasma instead of serum samples did generally not affect the levels of biomarkers resulting in a median recovery of 110.8% [range: 60.7% (HGF)-272% (OPN)]. Here, four biomarkers failed the range of recovery (Figure 5).
Launching the multiplex magnetic bead assay into clinical routine and thus providing an insight of diverse corporal processes by means of biomarkers would greatly support tumor diagnostics and differential diagnosis[20,21]. Here, an appropriate pre-analytical evaluation is indispensible in order to obtain reliable clinical results in the future[2,3].
As well-known for many research-use-only (RUO)-assays, there often is only limited data available concerning the methodological performance despite being distributed and used for study and research purposes[20-22]. This is quite relevant for the scientific community as the same names of the parameters are used in these compound tests as in regular in-vitro diagnostic (IVD) labeled assays which are commonly used in clinical laboratory routine diagnostics for which the methodical quality and clinical validity has been investigated thoroughly in most cases.
Certainly, the assay of the present study has to be perceived as a complex method consisting of as many different tests as markers are included. It seems to be quite challenging for manufacturers to optimize all markers in a multiparametric platform and to avoid interactions between them. This is all the more obvious by the conspicuous variety of the measured biomarkers regarding their chemical and physical characteristics. Thus, to evaluate the measurement quality of this assay the single markers have to be interpreted independently.
All in all, ten plates were run. The first five kits were ordered in one batch and applied subsequently under strictly identical conditions. The following plates were ordered and run six months later under the same standards of procedure. However, we perceived a striking discrepancy when comparing the results of the first five and the last five plates. As the lot number was the same, we assume an influence of surrounding conditions, e.g., room temperature, pipettes used and different laboratory staff constellation. In order to represent accurate and relevant results in our main method evaluation, we divided our evaluation into two steps: First, we considered only the first five assays as they were done under homogeneous conditions. In the second step, an overall analysis of all assays was performed. Nevertheless, this critical fact elucidates once more the importance of standardized procedures in clinical routine laboratories, the application of internal quality controls and the participation in external quality assessment programs[3,18].
In general, the assay showed an acceptable intra- and inter-assay imprecision. Apart from the synthetic internal controls QC 1 and QC 2 we included the physiological external control samples pool 1 and pool 2 as a further reference source. In comparison to the internal controls QC 1 and QC 2 this resulted in an interesting finding. As expected, the method precision was slightly more accurate for the synthetic samples. Although evidently the composition of serum pools is more complex, this physiological type of quality control is more relevant as it reflects the situation of the clinical samples more accurately. Nevertheless, the comparability between synthetic and physiological samples was still given. However, we perceived higher variation in samples with lower concentrations of the biomarkers, particularly seen in the imprecision results of pool 2 where most marker concentrations were below or at the lower end of the accepted range.
Comparing the precision data provided by the manufacturer (range: 4.9% to 15.0% and 4.1% to 16.2% for intra- and inter-assay imprecision, respectively) with our results a very good accordance concerning the intra-assay results was observed.
The inter-assay imprecision is partly higher in our tests. However, for most markers we could achieve the corresponding coefficients of variation considering the results of the quality controls provided by the manufacturer. CYFRA 21-1, a highly valuable tumor marker for non small cell lung cancer[16,23,24], was found to be the only marker exceeding the 20% limit of CV concurring with the given imprecision by the manufacturer where it also yields the maximum coefficient of variation in the panel. As the marker concentration is within the appropriate range and the variation is found to be not acceptable we assume a non-applicability of this biomarker in the assessed method. In our higher concentrated pool 1, five markers were found to be measured with a non-acceptable CV. Close observation of these biomarkers revealed that the observed concentrations were very low, even below the first standard (such as for FGF2) indicating only limited clinical relevance in these cases. In conclusion, with an inter-assay imprecision in a reasonable range between 4.59% (total PSA)-23.88% (TGFα), this is a rather satisfying result.
Comparing the values of variance of fluorescence intensity and observed concentration revealed some striking discrepancies. This phenomenon is obviously based on the transformation of measured fluorescence intensity into biomarker concentration, which is neither linear nor predetermined to be equal in all the tests, but corresponding to the course of the standard curve calculated anew for every single test. FI values are matched to the concentrations that are provided by the standard curve. Thus, the same FI values in two different plates could lead to two different concentrations and are therefore less comparable when it comes to a crossover comparison between the plates. This phenomenon is shown in the attached Table 3 comparing FI values and observed concentration-based CVs. Hence, the inter-assay imprecision based on concentration values provided a more relevant result for the methodological evaluation.
Testing the dilution linearity, our study yields satisfying results with a tendency to a recovery in the upper field of the defined acceptable range.
Within the Bio-Plex® 200 System, it is possible to vary the minimal number of events needed for measurement. With 50 events as minimum per bead we chose a compromise between a time-effective measurements and sufficiently precise results. Obviously, the accuracy of measurements increases with higher number of events. Nevertheless, our findings could achieve acceptable CV values in most cases despite the low minimum number of events set. Therefore, we did not compare absolute number of detected events and their calculated CVs.
The MILLIPLEX® MAP Human Circulating Cancer Biomarker Magnetic Bead Panel 1 offers a wide spectrum of applicable biomarkers. However, it is obviously neither clinically relevant nor cost-effective to apply the complete panel in diagnostics. After definition of the relevant markers for each tumor type, the panel must be focused and applied as a biomarker pattern of clinical interest depending on the contemplated entity of disease.
For implementation into clinical diagnostics, further studies evaluating its performance in a large cohort of cancer patients and appropriate control groups, which are relevant for differential diagnosis, i.e., healthy individuals and patients with organ related benign disease, are definitely required. Currently, we are performing such clinical validation studies with cohorts of patients suffering from gastrointestinal, gynecological and urological cancers.
Apart from the method quality itself, preanalytical handling of samples prior to analysis in the laboratory can influence the final results, as is known for several research and routine parameters[25-27]. Hence, we examined the stability of the tested markers. As often observed in the clinical routine, samples are not directly transferred to the central laboratory and instead remain exposed to room temperature without centrifugation. In order to depict this highly relevant situation, samples were centrifuged after 6 and 24 h. However, most markers remained stable, except MIF and IL-8. These two markers presented a considerable increase in marker levels. Our findings for IL-8 agree with the recommendations made by Hoch et al[28] to centrifuge the blood samples within less than 2 h to avoid interactions between IL-8 and blood cells. An increase of MIF levels in samples, which were not directly prepared, is also predescribed by Sobierajski et al[29]. These pre-analytical facts must be observed by the clinicians as prolonged storage before processing could lead to fatal misinterpretation in these markers.
Sample storage up to 48 h after centrifugation at a temperature of 4 °C showed a good outcome for all markers except OPN, which presented a decline after 6 h storage.
Storage at 25 °C after centrifugation showed a stronger effect on marker levels. While stability is given until 24 h, a more or less increase of marker concentrations can be observed after longer storage time. For example, the recovery of HE4 rises over 200% after 48 h. Again, OPN is the only marker showing a decline to nearly 80% in recovery supporting our above mentioned findings. Also, Cristaudo et al[30] found an instable performance of OPN after storage of serum samples at room temperature.
Furthermore, we observed good comparability between serum and EDTA-plasma samples. Only OPN and leptin-recoveries exceeded the 130% mark of the accepted range. These results correspond with the findings of Lanteri et al[31] and Gröschl et al[32] where an increase of these biomarkers in plasma samples compared with serum samples was observed. HGF presented a recovery of 60.7% as the unique marker undergoing the accepted 70% mark, also concurring with previous HGF stability analysis[33].
In our study, the evaluated method is demonstrated to be a stable and precise tool for detection of most biomarkers included in the kit. Unfortunately, for CYFRA 21-1, the method did not achieve acceptable inter-assay precision values. This should be further investigated. In general, we recommend that “research use only”-tests are assessed before implementation into further research and clinical routine. Here, certain preconditions, such as ordering tests in a batch, use of physiological quality controls in addition to the provided control samples as well as relevant pre-analytical aspects of some markers, should be observed.
All in all, this study shows that the MILLIPLEX® MAP Human Circulating Cancer Biomarker Magnetic Bead Panel 1 could offer new diagnostic perspectives while further studies are necessary to show its clinical applicability, usefulness and comparability with established routine assays.
We are grateful for access to the Biorad and Luminex instruments of the laboratory of Professor Knolle and the laboratory of Professor Hornung at the University Hospital Bonn, where the analyses were performed. This work is part of the doctoral theses of Hermann N and Dreßen K.
Cancer is a global health problem resulting in about eight million deaths each year. Tumor markers as additional diagnostic tools, which are easily assessable in blood, have been launched in the last century and with them a large variety of detecting methods. Of these, enzyme-linked immunosorbent assays are the standard methods in clinical routine today, while multiplexing has become interesting for cancer research as it is a quick, cheap, less-volume-wasting, easy-to-handle but still precise tool for parallel measurement of multiple markers. This goes hand in hand with the fact that nowadays cancer is perceived as a complex disease involving multiple processes. The authors investigated the MILLIPLEX® MAP Human Circulating Cancer Biomarker Magnetic Bead Panel 1, purchased from Millipore, which was specially designed for cancer diagnostics, on its methodical performance.
Most striking is the fact that many different assays are used for research purpose, although they are not that strictly proven as established methods for in-vitro diagnostics. However, research results are assigned to a clinical setting. Therefore, a thorough methodical investigation using physiological samples has to be performed.
Previous investigations of the multiplex technology used by Millipore showed good correlations to single ELISAs of each tested biomarker. However, here only a few markers were tested in parallel. Furthermore, this specially designed kit with the possibility to measure 24 biomarkers from the areas of angiogenesis, immunology, apoptosis as well as established and auspicious tumor markers has not been tested on its entirety regarding methodological performance and stability. In the investigations, the authors used serum pools as physiological control samples and most markers showed good results.
This study allows other users to assess the quality and applicability of the assay and to conduct further clinical studies on methodically solid bedrock.
All substances that can be measured in cancer patients and which reveal a malignant disease or contribute to its prognosis or treatment are called tumor markers. Multiplexing is one of the detection methods for tumor markers which are assessed in body fluids and it allows the parallel measurement of multiple markers. An ELISA is an enzyme-linked immunosorbent assay which is another detection method for tumor markers but limited to the measurement of only one marker per test. Both are antibody-based detection procedures and enzymatic color-reactions are used to quantify the results.
The manuscript is very well presented and highlights factors influencing the measurements of the key performance indicators of the multiplex cancer biomarker panel, which constitutes a highly interesting study. The findings from the comparison of the critical measurement parameters between physiological sera in parallel with synthetic internal controls will be of particular interest to a wide audience. The technical details are clearly defined and the interpretations are thorough with robust scientific conclusions.
P- Reviewer: Albulescu R, Das UN, Ferroni P, Liu H, Pang S, Tanase CP S- Editor: Song XX L- Editor: A E- Editor: Liu SQ
| 1. | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8406] [Cited by in RCA: 8971] [Article Influence: 690.1] [Reference Citation Analysis (0)] |
| 2. | Bretthauer M, Kalager M. Principles, effectiveness and caveats in screening for cancer. Br J Surg. 2013;100:55-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 3. | Sturgeon CM, Hoffman BR, Chan DW, Ch’ng SL, Hammond E, Hayes DF, Liotta LA, Petricoin EF, Schmitt M, Semmes OJ. National Academy of Clinical Biochemistry Laboratory Medicine Practice Guidelines for use of tumor markers in clinical practice: quality requirements. Clin Chem. 2008;54:e1-e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 107] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 4. | Sturgeon CM, Duffy MJ, Hofmann BR, Lamerz R, Fritsche HA, Gaarenstroom K, Bonfrer J, Ecke TH, Grossman HB, Hayes P. National Academy of Clinical Biochemistry Laboratory Medicine Practice Guidelines for use of tumor markers in liver, bladder, cervical, and gastric cancers. Clin Chem. 2010;56:e1-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 139] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 5. | Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313-5327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1111] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 6. | Holdenrieder S, Stieber P. Circulating apoptotic markers in the management of non-small cell lung cancer. Cancer Biomark. 2010;6:197-210. [PubMed] |
| 7. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47162] [Article Influence: 3368.7] [Reference Citation Analysis (5)] |
| 8. | Kellar KL, Douglass JP. Multiplexed microsphere-based flow cytometric immunoassays for human cytokines. J Immunol Methods. 2003;279:277-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 71] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Chowdhury F, Williams A, Johnson P. Validation and comparison of two multiplex technologies, Luminex and Mesoscale Discovery, for human cytokine profiling. J Immunol Methods. 2009;340:55-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 211] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 10. | Kim YW, Bae SM, Lim H, Kim YJ, Ahn WS. Development of multiplexed bead-based immunoassays for the detection of early stage ovarian cancer using a combination of serum biomarkers. PLoS One. 2012;7:e44960. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 11. | Nolen BM, Lokshin AE. Biomarker testing for ovarian cancer: clinical utility of multiplex assays. Mol Diagn Ther. 2013;17:139-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Park HD, Kang ES, Kim JW, Lee KT, Lee KH, Park YS, Park JO, Lee J, Heo JS, Choi SH. Serum CA19-9, cathepsin D, and matrix metalloproteinase-7 as a diagnostic panel for pancreatic ductal adenocarcinoma. Proteomics. 2012;12:3590-3597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Krishhan VV, Khan IH, Luciw PA. Multiplexed microbead immunoassays by flow cytometry for molecular profiling: Basic concepts and proteomics applications. Crit Rev Biotechnol. 2009;29:29-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 14. | Ellington AA, Kullo IJ, Bailey KR, Klee GG. Antibody-based protein multiplex platforms: technical and operational challenges. Clin Chem. 2010;56:186-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 222] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 15. | Horvath AR, Kis E, Dobos E. Guidelines for the use of biomarkers: principles, processes and practical considerations. Scand J Clin Lab Invest Suppl. 2010;242:109-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Molina R, Holdenrieder S, Auge JM, Schalhorn A, Hatz R, Stieber P. Diagnostic relevance of circulating biomarkers in patients with lung cancer. Cancer Biomark. 2010;6:163-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Moore RG, Jabre-Raughley M, Brown AK, Robison KM, Miller MC, Allard WJ, Kurman RJ, Bast RC, Skates SJ. Comparison of a novel multiple marker assay vs the Risk of Malignancy Index for the prediction of epithelial ovarian cancer in patients with a pelvic mass. Am J Obstet Gynecol. 2010;203:228.e1-228.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 191] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 18. | Dancey JE, Dobbin KK, Groshen S, Jessup JM, Hruszkewycz AH, Koehler M, Parchment R, Ratain MJ, Shankar LK, Stadler WM. Guidelines for the development and incorporation of biomarker studies in early clinical trials of novel agents. Clin Cancer Res. 2010;16:1745-1755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 186] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 20. | Fu Q, Zhu J, Van Eyk JE. Comparison of multiplex immunoassay platforms. Clin Chem. 2010;56:314-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 21. | Bastarache JA, Koyama T, Wickersham NE, Mitchell DB, Mernaugh RL, Ware LB. Accuracy and reproducibility of a multiplex immunoassay platform: a validation study. J Immunol Methods. 2011;367:33-39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Köhler K, Seitz H. Validation processes of protein biomarkers in serum--a cross platform comparison. Sensors (Basel). 2012;12:12710-12728. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 23. | Barak V, Goike H, Panaretakis KW, Einarsson R. Clinical utility of cytokeratins as tumor markers. Clin Biochem. 2004;37:529-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 243] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 24. | Holdenrieder S, von Pawel J, Dankelmann E, Duell T, Faderl B, Markus A, Siakavara M, Wagner H, Feldmann K, Hoffmann H. Nucleosomes and CYFRA 21-1 indicate tumor response after one cycle of chemotherapy in recurrent non-small cell lung cancer. Lung Cancer. 2009;63:128-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 25. | Yoshimura T, Fujita K, Kawakami S, Takeda K, Chan S, Beligere G, Dowell B. Stability of pro-gastrin-releasing peptide in serum versus plasma. Tumour Biol. 2008;29:224-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Lehner J, Wittwer C, Fersching D, Siegele B, Holdenrieder S, Stoetzer OJ. Methodological and preanalytical evaluation of an HMGB1 immunoassay. Anticancer Res. 2012;32:2059-2062. [PubMed] |
| 27. | Holdenrieder S, Mueller S, Stieber P. Stability of nucleosomal DNA fragments in serum. Clin Chem. 2005;51:1026-1029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Hoch RC, Schraufstätter IU, Cochrane CG. In vivo, in vitro, and molecular aspects of interleukin-8 and the interleukin-8 receptors. J Lab Clin Med. 1996;128:134-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 99] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 29. | Sobierajski J, Hendgen-Cotta UB, Luedike P, Stock P, Rammos C, Meyer C, Kraemer S, Stoppe C, Bernhagen J, Kelm M. Assessment of macrophage migration inhibitory factor in humans: protocol for accurate and reproducible levels. Free Radic Biol Med. 2013;63:236-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 30. | Cristaudo A, Foddis R, Bonotti A, Simonini S, Vivaldi A, Guglielmi G, Ambrosino N, Canessa PA, Chella A, Lucchi M. Comparison between plasma and serum osteopontin levels: usefulness in diagnosis of epithelial malignant pleural mesothelioma. Int J Biol Markers. 2010;25:164-170. [PubMed] |
| 31. | Lanteri P, Lombardi G, Colombini A, Grasso D, Banfi G. Stability of osteopontin in plasma and serum. Clin Chem Lab Med. 2012;50:1979-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 32. | Gröschl M, Wagner R, Dörr HG, Blum W, Rascher W, Dötsch J. Variability of leptin values measured from different sample matrices. Horm Res. 2000;54:26-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 33. | Nayeri F, Brudin L, Nilsson I, Forsberg P. Sample handling and stability of hepatocyte growth factor in blood samples. Cytokine. 2002;19:201-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |