Published online Sep 20, 2025. doi: 10.5662/wjm.v15.i3.98201
Revised: November 19, 2024
Accepted: December 23, 2024
Published online: September 20, 2025
Processing time: 259 Days and 2.3 Hours
Recently, the identification of cell apoptosis induced by natural products has become research hotspot and frontier in the biopharmaceutical and food indu
To establish a selective, instant, and practical protocol to identify cell apoptosis induced by natural products.
A one transient cell processing procedure (OTCPP) was used to detect human colorectal cancer LoVo cell apoptosis after treatment with Pinus massoniana bark extract (PMBE) at the morphological, biochemical, and cell cycle levels. The methods used included treatment with DNA gel electrophoresis, fluorescence microscopy, and flow cytometry.
In PMBE-treated LoVo cells, we observed a DNA ladder on gel electrophoresis and fluorescence microscopy revealed "nuclear shrinkage, chromatin conden
OTCPP can quickly identify apoptosis and measure the apoptosis rate, thereby unifying qualitative and quantitative analysis.
Core Tip: According to the global green development hypothesis, the identification of cell apoptosis induced by natural products is a research hotspot. However, routine laboratory identification of apoptosis suffers from experimental errors and poor repeatability. Herein, we explored the simultaneous morphological, biochemical and molecular identification of apoptosis, while reducing experimental errors. We established a protocol to instantly identify apoptosis following natural product treatment, termed one transient cell processing procedure (OTCPP), comprising eleven steps. It could identify apoptosis of cultured LoVo cells in a little as 4 days. The OTCPP procedure is easy, rapid and high efficient, especially for researchers in developing countries.
- Citation: Cui YY. Selective procedure for the instant identification of cellular apoptosis induced by natural products. World J Methodol 2025; 15(3): 98201
- URL: https://www.wjgnet.com/2222-0682/full/v15/i3/98201.htm
- DOI: https://dx.doi.org/10.5662/wjm.v15.i3.98201
Apoptosis is a normal physiological phenomenon[1,2], widely involved in development, tissue homeostasis, and some pathological processes, further becomes a frontier of bio-medical research. Classic methods for apoptosis identification include fluorescence microscopy, gel electrophoresis, and flow cytometry, among which DNA ladder resulting from gel electrophoresis is regarded as gold standard. However, these methods have different protocols and are performed with different cells cultured in different dishes, respectively, consuming more time. Apoptotic cells, whose degraded smaller DNA leaked out, fixed with ethanol and then stained with fluorescein, will become sub-diploids because of their DNA contents less than those of G1 phase[3,4]. The extracted DNA can be effectively controlled by phosphate-citric acid buffer (0.2 M, pH7.8), and the leaked DNA can form DNA ladder under gel electrophoresis, while the residual cells’ cell cycle distribution can be analyzed with flow cytometry[5,6].
Currently, the global green development hypothesis is promoting research into natural products, thus they have become hotspot in the biopharmaceutical and food industries. Certain natural products can promote or inhibit cell growth, or even induce their apoptosis[7]. The identification of natural product-induced apoptosis is thus an important subject. Consequently, the present study aimed to identify the effect of Pinus massoniana bark extract (PMBE) on human colorectal cancer LoVo cells to demonstrate the integrative procedure termed one transient cell processing procedure (OTCPP). OTCPP produces uses different instruments to achieve apoptosis detection, with relatively satisfactory results. We believe that OTCPP will play an important role in accelerating the progress of basic research, clinical diagnosis, and screening candidate natural drugs.
The Institute of Songzhen nutritional resource (Guangzhou, China) supplied the PMBE. The other reagents comprised HEPES (Amresco, Solon, OH, United States), Roswell Park Memorial Institute (RPMI) 1640 (HyClone, Logan, UT, United States), Penicillin-Streptomycin (Penicillin 10000 units/mL, Streptomycin 10000 µg/mL) (Invitrogen, Waltham, MA, United States), Fetal bovine serum (FBS; Sijiqing Co. Hangzhou, China), Trypsin (HyClone), propidium iodide (PI) (Sigma, St. Louis, MO, United States), RNase A (Sino-American Biotechnology Co., Beijing, China) and Proteinase K (Merck, Darmstadt).
This study used the human colorectal cancer cell line LoVo (ATCC, Manassas, VA, United States; number CCL-229).
The OTCPP process used an incubator, fluorescence microscope (BH-2, Olympus, Tokyo, Japan), electrophoresis apparatus (Bio-Rad, Hercules, CA, United States), a flow cytometer (ELITE, Beckman Coulter, Indianapolis, IN, United States), and MULTICYCLE analysis software (Phoenix Flow Systems, San Diego, CA, United States).
(1) Cell Culture and Treatment: Two wells of LoVo cells were cultured in complete RPMI 1640 media (10% FBS, 10 mmol/L HEPES, 100 IU/mL penicillin, 100 µg/mL streptomycin) in a flat-bottom plate (Costar, Corning, NY, United States), respectively, until to the logarithmic growth period (about 104-6 cells/mL), then labeled as experimental group and control group, further adding 140 µg/mL PMBE to the culture media or not, respectively, all incubating for 24 hours; (2) Cell Collection and fixation: 1 mL of 0.25% trypsin was used to trypsinize the LoVo cells (1-2 × 107) at 37℃ for appropriate time. Then, add 0.5% FBSRPMI 1640 media to stop trypsinization and gentle pipetting the cell clumps into single cells. The cells were transferred into an Eppendorf tube, centrifuged at 100 g for 5 minutes, and then discarded the supernatant. The cell pellet was washed twice using 1 × PBS with centrifugation at 100 g for 5 minutes each time. The cells were resuspended into single cell status using 1 × PBS (20 µL), added with 2 mL 70% ethanol, and placed at -20℃ overnight to fix the cells; (3) Centrifuged 100 g, 10 minutes, discarded supernatant to remove ethanol; (4) Resuspended in 0.5 mL 1 × PBS, transferred to a new micro-tube, centrifuged 150 g, 10 minutes, discarded supernatant; (5) Added 40 µL of 0.2 M phosphate-citric acid buffer (pH7.8), RT 30 minutes, intermittent shaking; (6) Centrifuged 100 g, 10 minutes, and then transferred the supernatant to a new micro-tube, incubated on ice for fluorescence observation and flow cytometry analysis; (7) Added 3 µL of 0.25% NP40, 3 µL of RNase A solution (1 mg/mL) to the supernatant, vortexed, further incubated at 37℃, 30 minutes; (8) Added 3 µL of Proteinase K (1 mg/mL) to the mixture, vortexed thoroughly, and then incubated at 37℃ for 30 minutes; (9) Then, 16 µL of the mixture was mixed thoroughly with 2 µL of 6 × DNA loading buffer, resolved on agarose gel at 220V, 2 hours, and then stained with EB. The gel was visualized under UV light; (10) Cell pellets of step (6) were resuspended in 0.5 mL 1 × PBS. Then, added 10 µL of Proteinase K (1 mg/mL), vortexed gently, and left RT, 30 minutes, then washed the cells two times using 1 × PBS with centrifugation 100 g, 5 minutes each time, and discarded supernatant, resuspended again in 20 µL 1 × PBS into single cells. Then, added 300 µL of DNA staining solution (containing 150 µg/mL PI and 20 U/mL RNase A) and incubated RT, 30 minutes; and (11) Placed a drop of the single cell suspension on a air-drying glass slide, covered with a coverslip, and observed under a fluorescence microscope (G.B filter, 400 ×). The remaining cell suspension for flow cytometry, analyzing the ratio of apoptotic cells.
4 days (2 ON incubation, 1 ON fixation, and 1 day with 2-3 hours of operation time).
(1) Trypsinize the cells just well; (2) Rotation speed no more than 150 g; (3) Resuspend the cells into a single cell suspension using a small amount of 1 × PBS, then fix them with ethanol; and (4) Recommend removing large multi-cellular aggregates with a nylon monofilament mesh screen before analysis on the flow cytometer, ensuring that the cells flow through the cytometer in a single line.
Using a one-station operation for one-time cell culture and OTCPP, obtained enough experimental materials to identify cell apoptosis with different instruments, letting synchronous identification of LoVo cell apoptosis at morphological, biochemical, cell cycle, and DNA content level, respectively, come true. Further, reach the perfect combination of qualitative and quantitative analysis, which not only shortened the experimental time, but also completed the task in four working days rather than the original 9 working days. The results were quite satisfactory (Figure 1, Figure 2, and Figure 3). Briefly, an integrative protocol for apoptosis identification and measurement was established.
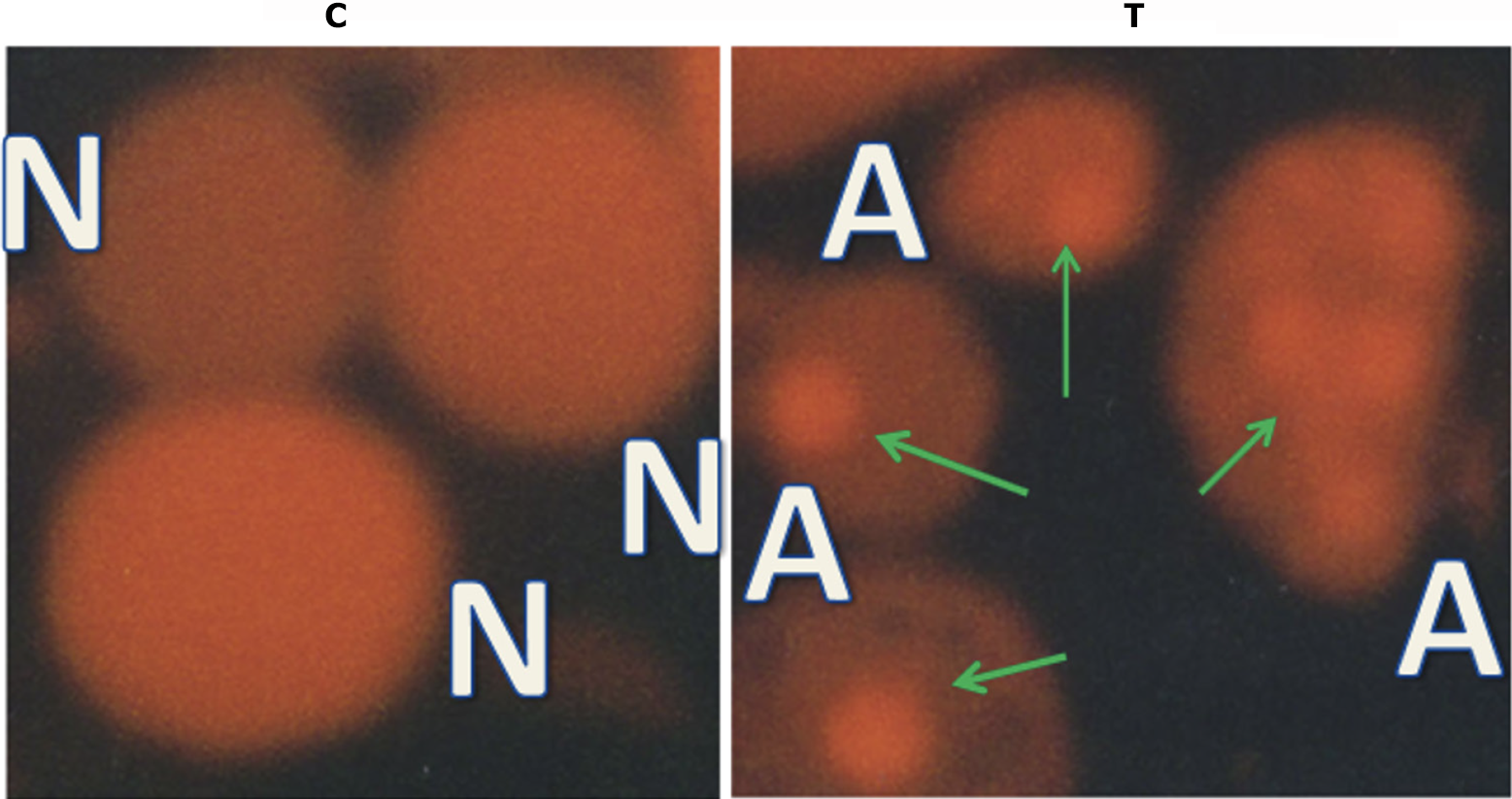
PI staining of control LoVo cells and cells treated with 140 µg/mL PMBE for 24 hours under a fluorescence microscope with a G.B. filter. Normal cells (N) depicted in the panel show light and homogeneous staining of their nuclei. In contrast, apoptotic cells (A) in the panel show thick and irregular staining of their nuclei. Magnification, 400 ×.
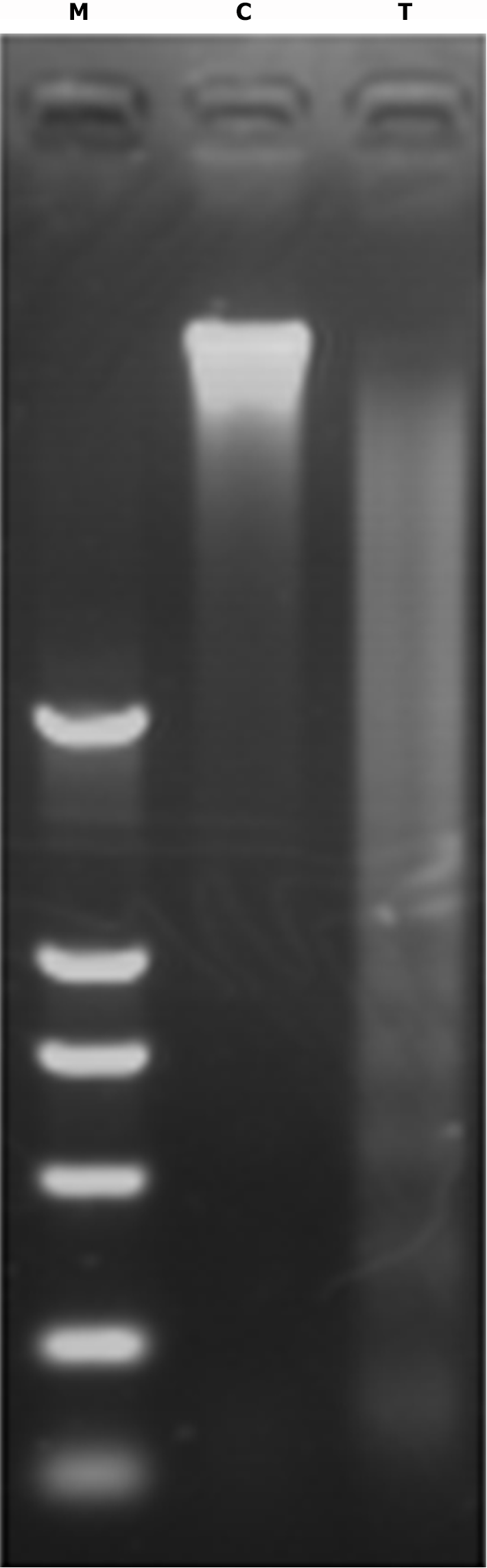
DNA ladder from apoptotic LoVo cells treated with 140 µg/mL PMBE for 24 hours. Results of gel electrophoresis showing that relatively low molecular-weight DNA had extruded out of apoptotic LoVo cells. M DNA marker DL2000; 1 Control; 2 PMBE Treatment
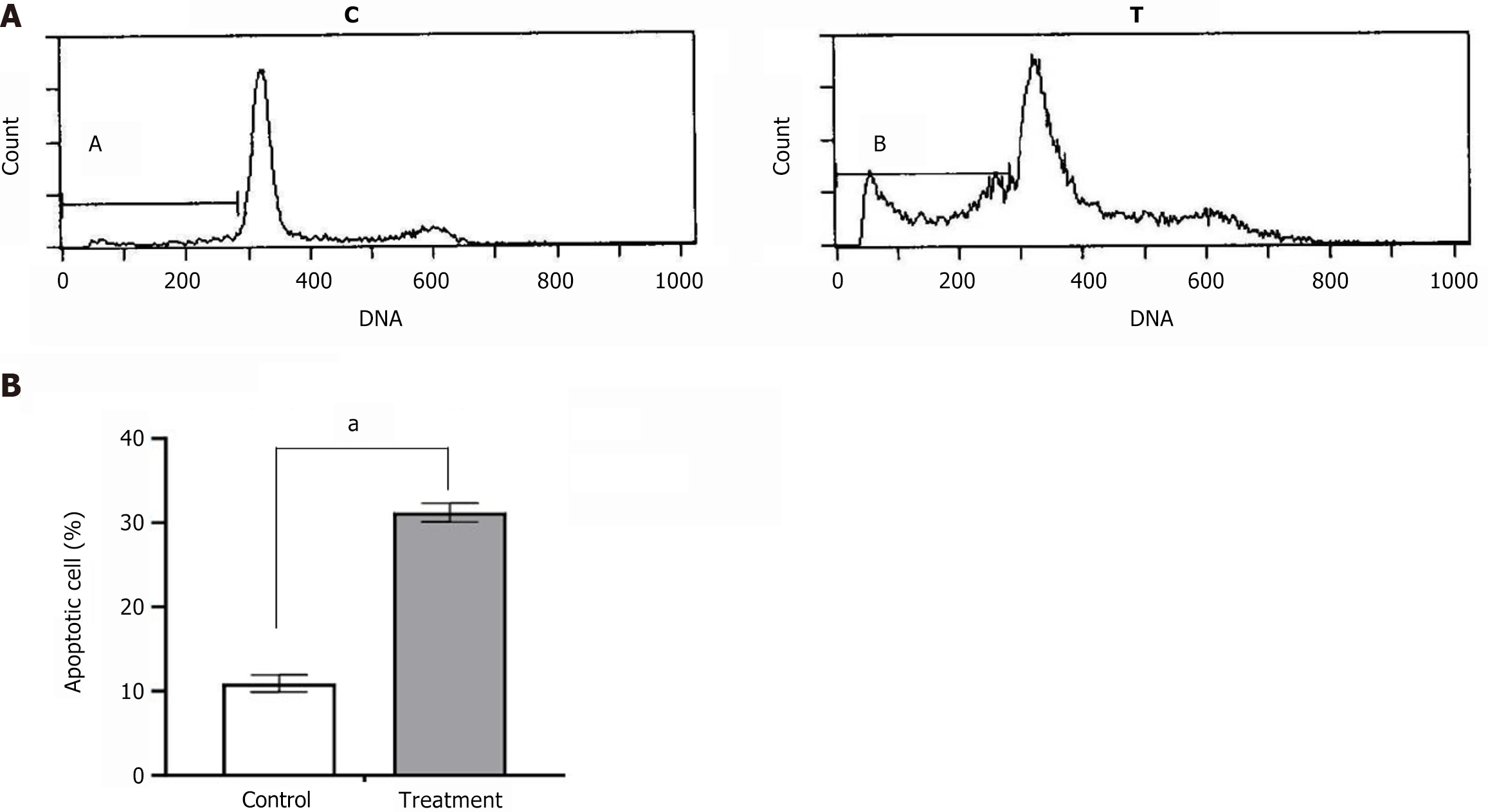
Results of flow cytometry analysis of control LoVo cells and LoVo cells treated with 140 µg/mL PMBE for 24 hours: (1) Shows a normal cell cycle distribution of untreated LoVo cells stained with PI, showing no obvious apoptotic-sub-G1 curve, but with 10.9% of the cells showing spontaneous apoptosis; (2) Shows the cell cycle distribution of LoVo cell treated with PMBE and stained with PI. There is an obvious apoptotic-sub-G1 curve before the G1 curve, with 31.4% of the cells showing apoptosis; and (3) Shows the statistical histogram of LoVo cell apoptosis induced by PMBE treatment compared with Control. aP < 0.05 vs Control, n = 3, mean ± SD.
Through culturing cells once and subsequent OCTPP, obtain enough experimental materials to detect the target cell apoptosis with different instruments, reaching the simultaneous identification of apoptosis at morphological, biochemical, cell cycle, and DNA content level, respectively, further realize the unity of quantitative and qualitative analyses, shortening the experimental time obviously, while producing relatively good results.
Current measures for apoptosis identification involve detecting morphological, biochemical, and DNA content changes. At morphological level, fluorescence microscopy requires culturing cells on coverslips until monolayers form, which involves in the complicated steps and the inferior picture quality due to cell-cell junctions. What’s more, it is qualitative other than quantitative. At molecular level, DNA for gel electrophoresis was not extracted classically after cells are lysed with buffer, not only time-consuming, but also potentially toxic, and possibly shearing DNA as well, further decreasing the reproducibility of experiments. Although pulse alternative field gel electrophoresis can avoid shearing DNA via pre-embedding cells in low-melting-point agarose, which will increase the time taken and the cost of the experiments. Moreover, these DNA detection methods are qualitative, or at most semi-quantitative. By contrast, flow cytometry is simple, rapid, quantitative, and multi-parametric.
Performing these three independent experiment results analyses (morphological, biochemical, and DNA content analyses) is time consuming, and the experimental materials are frequently not from the same cell culture batch, making it hard to ensure the consistency and veracity of the results. Herein, prefixed the cells in 70% cold ethanol, then 0.2 M phosphate-citrate buffer at pH7.8 was used to extract DNA, and then spin 100 g, 5-10 minutes to make the smaller DNA leaked out thoroughly. The supernatant was then sequentially incubated with RNase A and proteinase K, further for direct electrophoresis. The cell pellets were stained with PI, a proportion of which was for slide preparation and fluorescence microscope observation, the remaining for flow cytometry. OTCPP allows the rapid identification of the classical hallmarks of apoptosis, which owns merits as follows: (1) 70% cold ethanol fixation makes cells stored for relatively longer time before analysis, without any significant DNA degradation from pathogenic infection and cell autolysis; (2) Multiple targets (nuclear shrinkage, DNA ladder and sub-G1 curve) can be detected almost simultaneously, shortening the experimental time sharply; (3) Culturing cells on coverslips for nuclear morphology observation can be omitted completely; (4) DNA extraction is simple and rapid, with no phenol, chloroform, or other toxic reagents, and the inconvenience of pulse field gel electrophoresis; (5) 0.2 M phosphatecitrate buffer (pH 7.8) helps the smaller DNA fragments completely extruded out of apoptotic cells[8], increasing the sensitivity and specificity of flow cytometry; and (6) The same experimental samples can be ensured, enhancing the consistency during experimentation. Briefly and vividly, OTCPP successfully achieved three birds of cell apoptosis in one stone. Thus, OTCPP is an improvement on classical methods of apoptosis identification, and researchers can select it according to their different purposes and own experimental conditions. However, OTCPP might have potential limitations, unsuitable for long-lived cells lacking or just weak proliferative ability, e.g. neural cells, myocardial cells, and stem cells with relatively small numbers in different tissues in organisms.
The present study presents a novel selective procedure for the rapid identification of human colon cancer LoVo cell apoptosis induced by the natural product, PMBE, using the OTCPP protocol. The use of OTCPP might pave the way for future basic and clinical research by optimizing experimental conditions (e.g. single-molecular augmented capture and imaging) to improve detection sensitivity in other cell types and/or disease models, e.g. neurons, cardiomyocytes and stem cells in critical diseases.
I would like to express my sincere gratitude to my laboratory mates for their invaluable assistance which significantly improved this work.
| 1. | Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239-257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9960] [Cited by in RCA: 10003] [Article Influence: 188.7] [Reference Citation Analysis (0)] |
| 2. | Nössing C, Ryan KM. 50 years on and still very much alive: 'Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics'. Br J Cancer. 2023;128:426-431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 54] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 3. | Darzynkiewicz Z, Bruno S, Del Bino G, Gorczyca W, Hotz MA, Lassota P, Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1368] [Cited by in RCA: 1406] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 4. | Costigan A, Hollville E, Martin SJ. Discriminating Between Apoptosis, Necrosis, Necroptosis, and Ferroptosis by Microscopy and Flow Cytometry. Curr Protoc. 2023;3:e951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 5. | Hotz MA, Gong J, Traganos F, Darzynkiewicz Z. Flow cytometric detection of apoptosis: comparison of the assays of in situ DNA degradation and chromatin changes. Cytometry. 1994;15:237-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 175] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 6. | Gong J, Traganos F, Darzynkiewicz Z. A selective procedure for DNA extraction from apoptotic cells applicable for gel electrophoresis and flow cytometry. Anal Biochem. 1994;218:314-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 480] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 7. | Cui YY, Xie H, Wang JF. [Transient detection of apoptosis of human liver cancer cells induced by Pinus massoniana bark extract (PMBE) in vitro]. Zhongguo Zuzhihuaxue Yu Xibaohuaxue Zazhi. 2005;14:80-83. |
| 8. | Zhu XQ, Wang GS, Zhang XJ, Zhao Q. [Effects of sodium ferculate on the viability and apoptosis of human hepatoma BEL-7404 cells]. Shijie Huaren Xiaohua Zazhi. 1999;7:715-716. |